Detection of Emerging Pollutants Using Aptamer-Based Biosensors: Recent Advances, Challenges, and Outlook
Abstract
:1. Introduction
2. Fundamentals of Aptamer-Based Biosensors
2.1. Aptamer Development
2.2. Types of Aptasensors
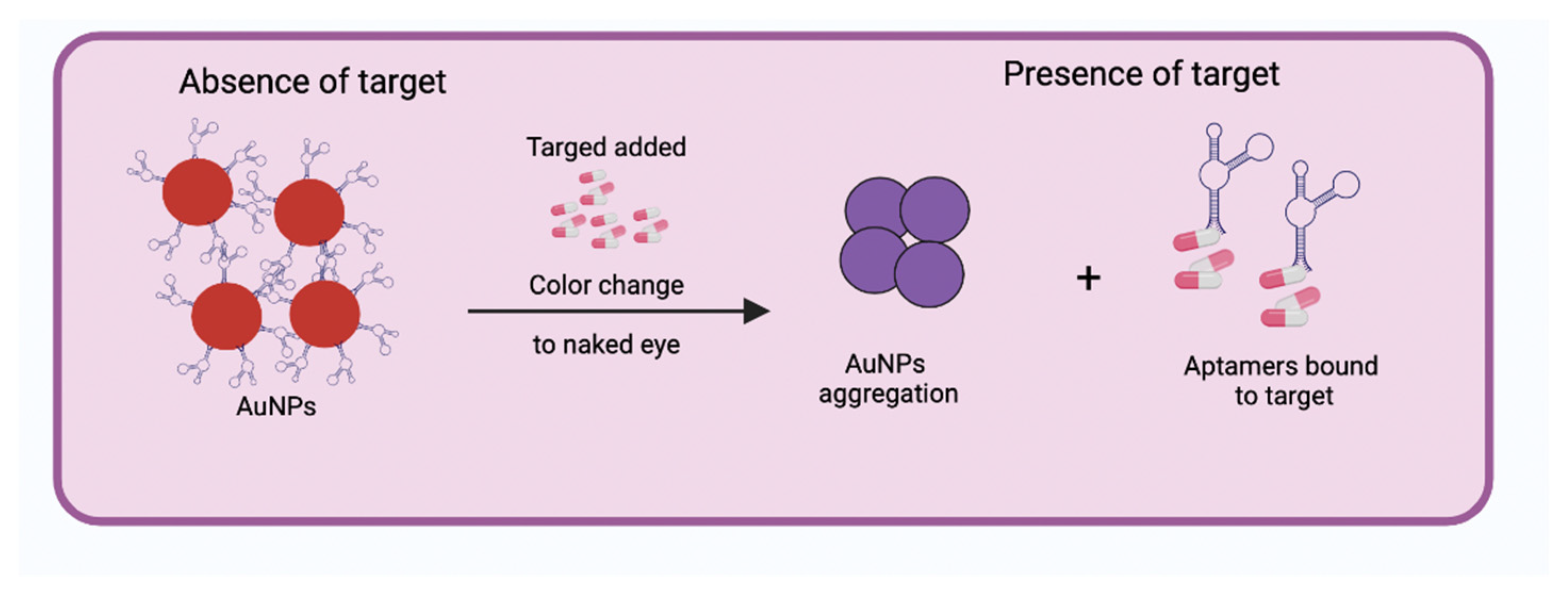
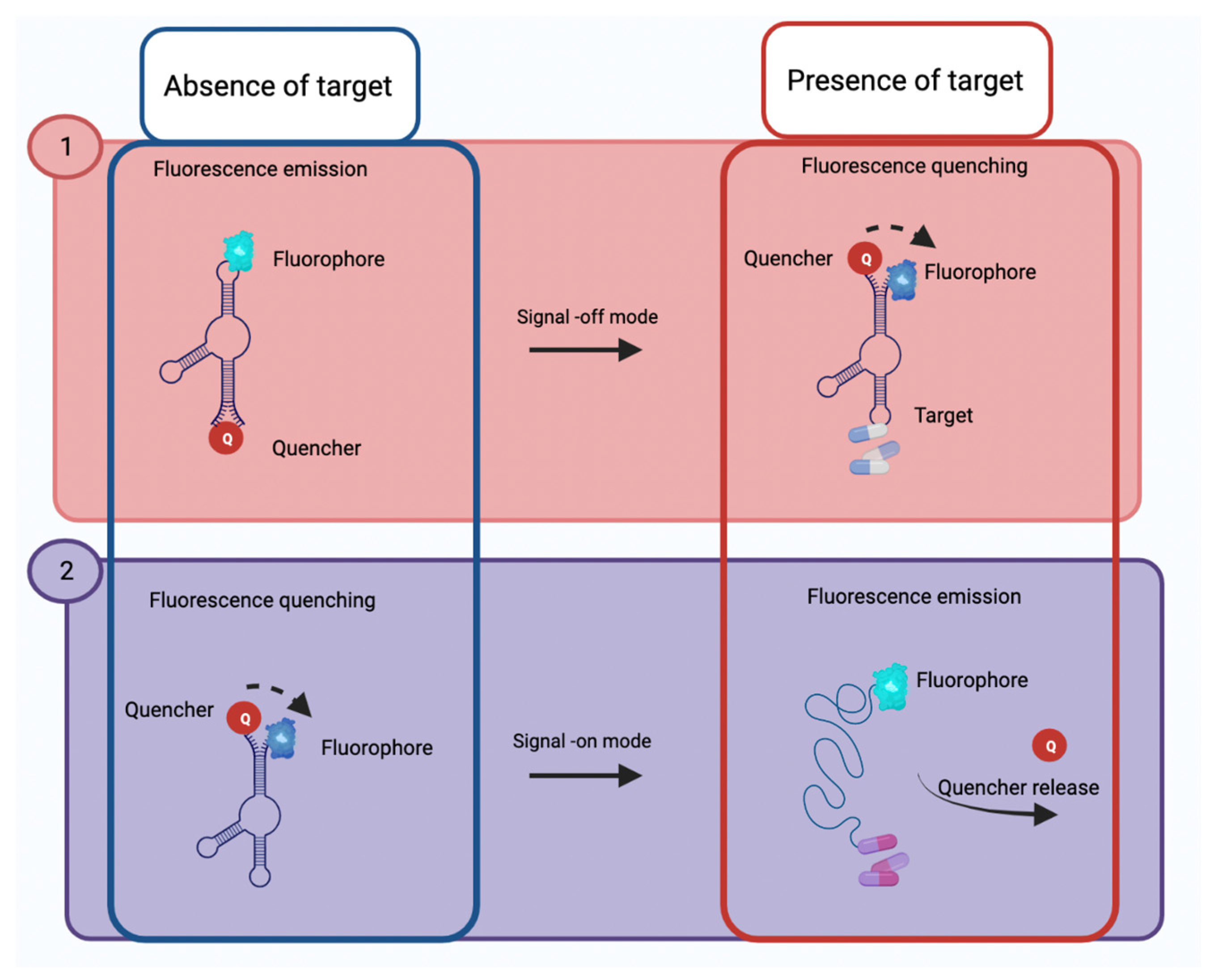
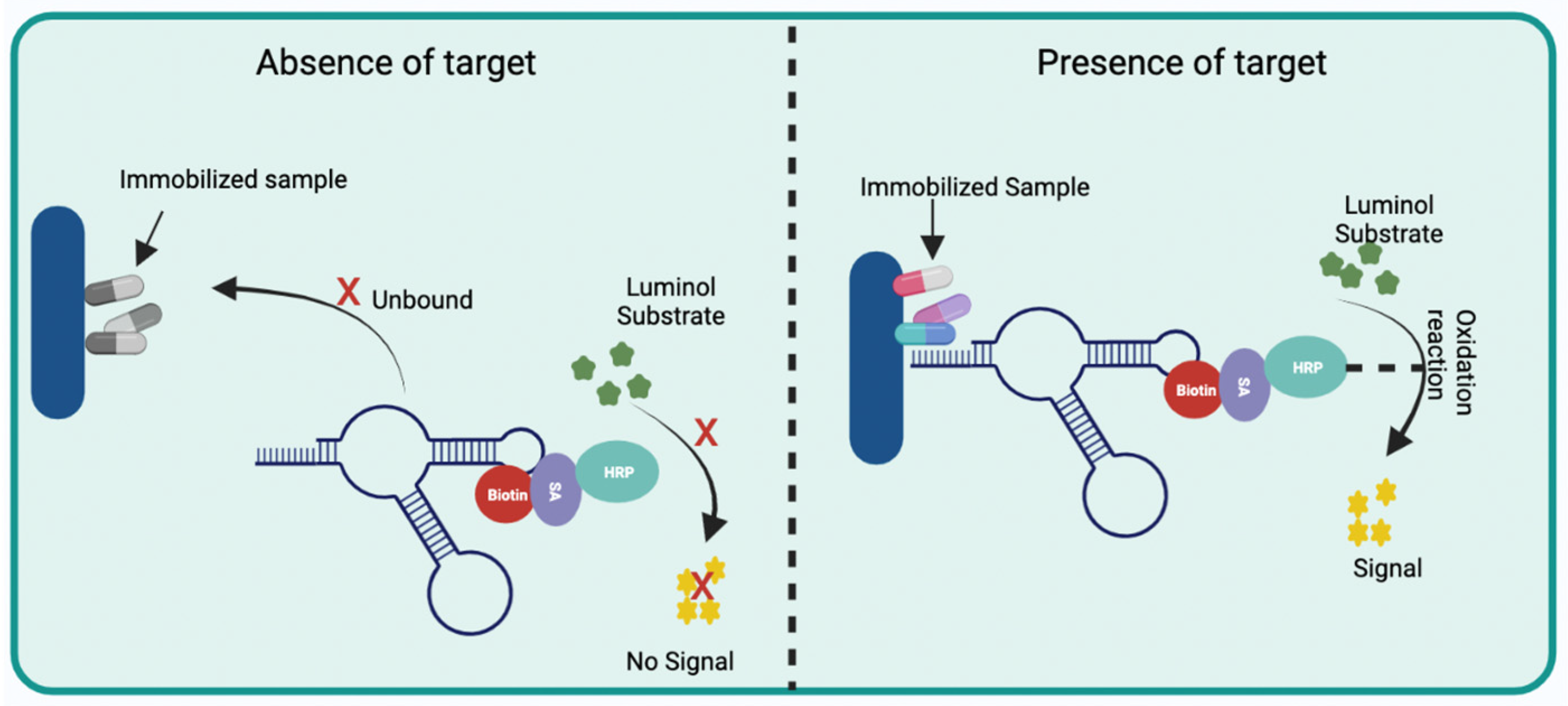
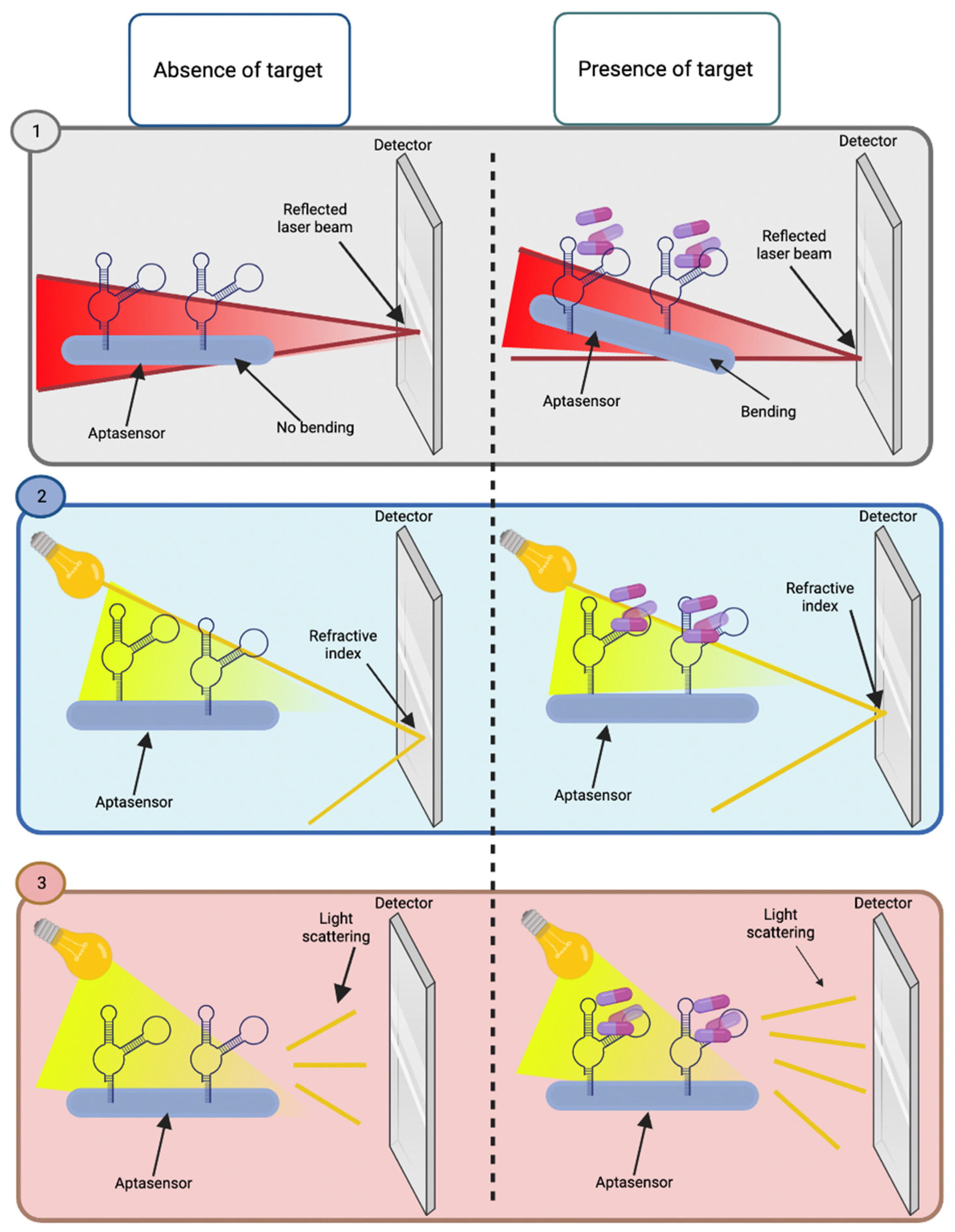
2.2.1. Optical Aptasensors
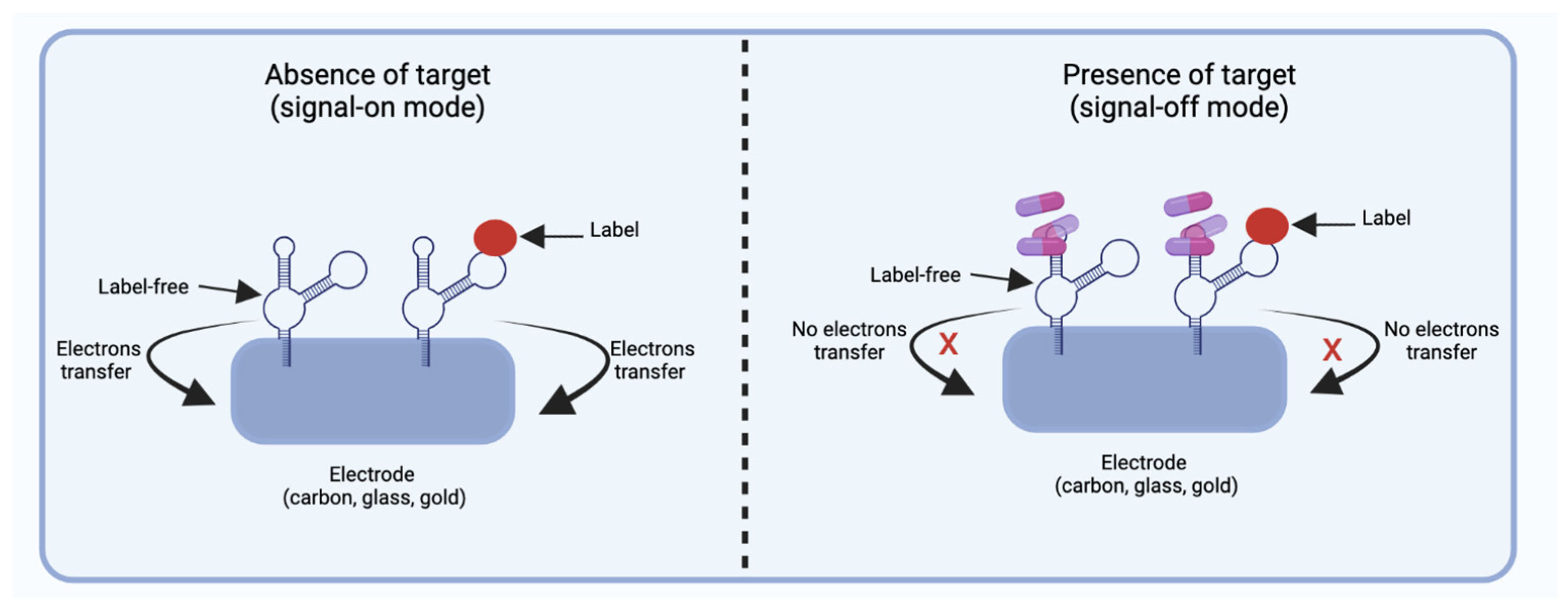
2.2.2. Electrochemical Aptasensors
2.2.3. Optics-Mass Aptasensors
3. Recent Advances in Aptamer-Based Biosensors
3.1. Biological Contaminants
3.2. Pharmaceutical and Personal Care Products
3.3. Endocrine-Disrupting Chemicals
3.4. Heavy Metals
3.5. Agricultural Compounds
4. Current Challenges and Recommendations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- González-González, R.B.; Flores-Contreras, E.A.; Parra-Saldívar, R.; Iqbal, H.M.N. Bio-removal of emerging pollutants by advanced bioremediation techniques. Environ. Res. 2022, 214, 113936. [Google Scholar] [CrossRef] [PubMed]
- Eissa, F.; Al-Sisi, M.; Ghanem, K. Occurrence and ecotoxicological risk assessment of pesticides in sediments of the Rosetta branch, Nile River, Egypt. J. Environ. Sci. 2022, 118, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Peris, A.; Barbieri, M.V.; Postigo, C.; Rambla-Alegre, M.; López de Alda, M.; Eljarrat, E. Pesticides in sediments of the Ebro River Delta cultivated area (NE Spain): Occurrence and risk assessment for aquatic organisms. Environ. Pollut. 2022, 305, 119239. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, D.; Nałęcz-Jawecki, G.; Drzewicz, P.; Skowronek, A.; Mianowicz, K.; Strzelecka, A.; Giebułtowicz, J. The assessment of environmental risk related to the occurrence of pharmaceuticals in bottom sediments of the Odra River estuary (SW Baltic Sea). Sci. Total Environ. 2022, 828, 154446. [Google Scholar] [CrossRef]
- Chacón, L.; Reyes, L.; Rivera-Montero, L.; Barrantes, K. Transport, fate, and bioavailability of emerging pollutants in soil, sediment, and wastewater treatment plants: Potential environmental impacts. In Emerging Contaminants in the Environment; Sarma, H., Dominguez, D.C., Lee, W.-Y., Eds.; Elsevier: Cambridge, MA, USA, 2022; pp. 111–136. [Google Scholar] [CrossRef]
- Albero, B.; Tadeo, J.L.; Pérez, R.A. Determination of Emerging Contaminants in Cereals by Gas Chromatography-Tandem Mass Spectrometry. Front. Chem. 2020, 8, 571668. [Google Scholar] [CrossRef]
- Shreenidhi, K.S.; Priyavadhana, P.; Purnima, N.; Rashminiza, A.; Sneha, S.; Vijaya Geetha, B. Study on the toxic effects of pharmaceutical drugs–Norfloxacin using Pangasius Sp. fish model and its mitigation using Artemisia pallens. Acta Ecol. Sin. 2022. [Google Scholar] [CrossRef]
- McCallum, E.S.; Sundelin, A.; Fick, J.; Alanärä, A.; Klaminder, J.; Hellström, G.; Brodin, T. Investigating tissue bioconcentration and the behavioural effects of two pharmaceutical pollutants on sea trout (Salmo trutta) in the laboratory and field. Aquat. Toxicol. 2019, 207, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Polverino, G.; Martin, J.M.; Bertram, M.G.; Wiles, S.C.; Palacios, M.M.; Bywater, C.L.; White, C.R.; Wong, B.B.M. Chronic exposure to a pervasive pharmaceutical pollutant erodes among-individual phenotypic variation in a fish. Environ. Pollut. 2020, 263, 114450. [Google Scholar] [CrossRef] [PubMed]
- Fenner, K.; Scheringer, M. The Need for Chemical Simplification As a Logical Consequence of Ever-Increasing Chemical Pollution. Environ. Sci. Technol. 2021, 55, 14470–14472. [Google Scholar] [CrossRef]
- Saldarriaga-Hernandez, S.; Hernandez-Vargas, G.; Iqbal, H.M.N.; Barceló, D.; Parra-Saldívar, R. Bioremediation potential of Sargassum sp. biomass to tackle pollution in coastal ecosystems: Circular economy approach. Sci. Total Environ. 2020, 715, 136978. [Google Scholar] [CrossRef]
- González-González, R.B.; Flores-Contreras, E.A.; González-González, E.; Castillo, N.E.T.; Parra-Saldívar, R.; Iqbal, H.M.N. Biosensor Constructs for the Monitoring of Persistent Emerging Pollutants in Environmental Matrices. Ind. Eng. Chem. Res. 2022. [Google Scholar] [CrossRef]
- Rizwan, K.; Rahdar, A.; Bilal, M.; Iqbal, H.M.N. MXene-based electrochemical and biosensing platforms to detect toxic elements and pesticides pollutants from environmental matrices. Chemosphere 2022, 291, 132820. [Google Scholar] [CrossRef] [PubMed]
- Nurani, S.; Abdul, H.; Lau, W. Detection of contaminants in water supply: A review on state-of-the-art monitoring technologies and their applications. Sens. Actuators B Chem. 2020, 255, 2657–2689. [Google Scholar]
- Phopin, K.; Tantimongcolwat, T. Pesticide Aptasensors—State of the Art and Perspectives. Sensors 2020, 20, 6809. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divya; Dkhar, D.S.; Kumari, R.; Mahapatra, S.; Kumar, R.; Chandra, P. Ultrasensitive Aptasensors for the Detection of Viruses Based on Opto-Electrochemical Readout Systems. Biosensors 2022, 12, 81. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Nguyen, J.; Li, Y. Aptamer-Based Biosensors for Environmental Monitoring. Front. Chem. 2020, 8, 434. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, L.; Mazloum-Ardakani, M. Advances in aptasensor technology. Adv. Clin. Chem. 2020, 99, 237–279. [Google Scholar] [CrossRef] [PubMed]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-Based Biosensors for Antibiotic Detection: A Review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Zhang, J. Protein Assay Based on Protein-Small Molecule Interaction; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 9780128150535. [Google Scholar]
- Mei, M.; Mu, L.; Liang, S.; Wang, Y.; She, G.; Shi, W. A general configurational strategy to quencher-free aptasensors. Biosens. Bioelectron. 2021, 178, 113025. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yan, X.; Yang, Y.; Qi, X.; Zhao, Y.; Li, L.; Ma, R.; Wang, L.; Dong, Y.; Sun, J.; et al. Advances and perspectives of aptasensors for the detection of tetracyclines: A class of model compounds of food analysis. Food Chem. 2021, 364, 130361. [Google Scholar] [CrossRef] [PubMed]
- Davydova, A.; Vorobyeva, M. Aptamer-Based Biosensors for the Colorimetric Detection of Blood Biomarkers: Paving the Way to Clinical Laboratory Testing. Biomedicines 2022, 10, 1606. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Jiang, X.; Li, N.; Jiang, X.; Liu, C.; Xu, J.J.; Wu, P. A Portable, Battery-Powered Photoelectrochemical Aptasesor for Field Environment Monitoring of E. coli O157:H7. Sens. Actuators B 2021, 346, 130520. [Google Scholar] [CrossRef]
- Ren, J.; Liang, G.; Man, Y.; Li, A.; Jin, X.; Liu, Q.; Pan, L. Aptamer-based fluorometric determination of Salmonella typhimurium using Fe3O4 magnetic separation and CdTe quantum dots. PLoS ONE 2019, 14, e0218325. [Google Scholar] [CrossRef]
- Lu, C.; Gao, X.; Chen, Y.; Ren, J.; Liu, C. Aptamer-Based Lateral Flow Test Strip for the Simultaneous Detection of Salmonella typhimurium, Escherichia coli O157:H7 and Staphylococcus aureus. Anal. Lett. 2020, 53, 646–659. [Google Scholar] [CrossRef]
- Peinetti, A.S.; Lake, R.J.; Cong, W.; Cooper, L.; Wu, Y.; Ma, Y.; Pawel, G.T.; Toimil-Molares, M.E.; Trautmann, C.; Rong, L.; et al. Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci. Adv. 2021, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Shi, A.; Zheng, Z.; Huang, L.; Wang, Y.; Lin, H.; Lin, X. Simultaneous quantification of ampicillin and kanamycin in water samples based on lateral flow aptasensor strip with an internal line. Molecules 2021, 26, 3806. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, C.; Sun, Y.; Shao, Y.; Cai, Y.; Zhang, Y.; Miao, J.; Miao, P. A colorimetric aptasensor for the antibiotics oxytetracycline and kanamycin based on the use of magnetic beads and gold nanoparticles. Microchim. Acta 2018, 185, 548. [Google Scholar] [CrossRef]
- Tu, D.; Garza, J.T.; Coté, G.L. A SERS aptasensor for sensitive and selective detection of bis(2-ethylhexyl) phthalate. RSC Adv. 2019, 9, 2618–2625. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, Z.; Liu, S.; Zhao, G. A highly sensitive and group-targeting aptasensor for total phthalate determination in the environment. J. Hazard. Mater. 2021, 412, 125174. [Google Scholar] [CrossRef]
- Wei, Y.; Zhou, Y.; Wei, Y.; Dong, C.; Wang, L. A fluorescent aptasensor based on berberine for ultrasensitive detection of bisphenol A in tap water. Anal. Methods 2021, 13, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; He, D.; Li, S.; Chen, R.; Peng, Y.; Li, S.; Han, D.; Wang, Y.; Qin, K.; Ren, S.; et al. Magnetic Halloysite Nanotube-Based SERS Biosensor Enhanced with Au@Ag Core-Shell Nanotags for Bisphenol A Determination. Biosensors 2022, 12, 387. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, D.; Liu, Y.; Zhan, X.; Lu, Y.; Zhou, P.; Zhang, D. An Electrochemical Aptasensor for Pb2+ Detection Based on Metal-Organic-Framework-Derived Hybrid Carbon. Biosensors 2020, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chen, C.; Guo, X.; Liu, C.; Yang, W. A simple electrochemiluminesecence aptasenor using a GCE/NCQDs/aptamers for detection of Pb. Environ. Technol. 2021, 43, 2270–2277. [Google Scholar] [CrossRef]
- Siddiqui, M.F.; Khan, Z.A.; Jeon, H.; Park, S. SPE based soil processing and aptasensor integrated detection system for rapid on site screening of arsenic contamination in soil. Ecotoxicol. Environ. Saf. 2020, 196, 110559. [Google Scholar] [CrossRef]
- Pan, J.; Li, Q.; Zhou, D.; Chen, J. Ultrasensitive aptamer biosensor for arsenic (III) detection based on label-free triple-helix molecular switch and fluorescence sensing platform. Talanta 2018, 189, 370–376. [Google Scholar] [CrossRef]
- Ensafi, A.A.; Akbarian, F.; Heydari-Soureshjani, E.; Rezaei, B. A novel aptasensor based on 3D-reduced graphene oxide modified gold nanoparticles for determination of arsenite. Biosens. Bioelectron. 2018, 122, 25–31. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Wang, Y.S.; Zhou, B.; Yu, J.H.; Peng, L.L.; Huang, Y.Q.; Li, X.J.; Chen, S.H.; Tang, X.; Wang, X.F. A multifunctional fluorescent aptamer probe for highly sensitive and selective detection of cadmium(II). Anal. Bioanal. Chem. 2017, 409, 4951–4958. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Nguyen, V.T.; Park, J.G.; Gu, M.B. Detection of iprobenfos and edifenphos using a new multi-aptasensor. Anal. Chim. Acta 2015, 868, 60–66. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, L.; Tu, Z.; Sun, X.; He, Q.; Lei, Z.; Xu, C.; Liu, Y.; Zhang, X.; Yang, J.; et al. Organophosphorus pesticides detection using broad-specific single-stranded DNA based fluorescence polarization aptamer assay. Biosens. Bioelectron. 2014, 55, 216–219. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Drinking Water Contaminant Candidate List 5; U.S. Environmental Protection Agency: Washington, DC, USA, 2021.
- Guo, L.; Wan, K.; Zhu, J.; Ye, C.; Chabi, K.; Yu, X. Detection and distribution of vbnc/viable pathogenic bacteria in full-scale drinking water treatment plants. J. Hazard. Mater. 2021, 406, 124335. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, S.V.; Vadde, K.K.; Phan, D.C.; Jafarzadeh, A.; Kapoor, V. Assessing the impact of flooding on bacterial community structure and occurrence of potentially pathogenic bacteria in Texas Rivers after Hurricane Harvey. J. Hazard. Mater. Lett. 2022, 3, 100058. [Google Scholar] [CrossRef]
- Felföldi, T.; Heéger, Z.; Vargha, M.; Márialigeti, K. Detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary. Clin. Microbiol. Infect. 2010, 16, 89–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, S.; Kitamura, G.; Segawa, T.; Kobayashi, A.; Miura, T.; Sano, D.; Okabe, S. Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl. Environ. Microbiol. 2014, 80, 7505–7511. [Google Scholar] [CrossRef] [Green Version]
- Zhan, L.; Li, C.M.; Fu, Z.F.; Zou, H.Y.; Huang, C.Z. Dual-aptamer-based enzyme linked plasmonic assay for pathogenic bacteria detection. Colloids Surf. B Biointerfaces 2022, 214, 112471. [Google Scholar] [CrossRef]
- Fang, S.; Song, D.; Zhuo, Y.; Chen, Y.; Zhu, A.; Long, F. Simultaneous and Sensitive Determination of Escherichia coli O157:H7 and Salmonella typhimurium Using Evanescent Wave Dual-Color Fluorescence Aptasensor Based on Micro/Nano Size Effect. Biosens. Bioelectron. 2021, 185, 113288. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014; Volume 30.
- González-González, R.B.; Sharma, A.; Parra-Saldívar, R.; Ramirez-Mendoza, R.A.; Bilal, M.; Iqbal, H.M.N. Decontamination of emerging pharmaceutical pollutants using carbon-dots as robust materials. J. Hazard. Mater. 2022, 423, 127145. [Google Scholar] [CrossRef]
- Lübbert, C.; Baars, C.; Dayakar, A.; Lippmann, N.; Rodloff, A.C.; Kinzig, M.; Sörgel, F. Environmental pollution with antimicrobial agents from bulk drug manufacturing industries in Hyderabad, South India, is associated with dissemination of extended-spectrum beta-lactamase and carbapenemase-producing pathogens. Infection 2017, 45, 479–491. [Google Scholar] [CrossRef]
- Xu, X.; Xu, Y.; Xu, N.; Pan, B.; Ni, J. Pharmaceuticals and personal care products (PPCPs) in water, sediment and freshwater mollusks of the Dongting Lake downstream the Three Gorges Dam. Chemosphere 2022, 301, 134721. [Google Scholar] [CrossRef]
- Guedes-Alonso, R.; Sosa-Ferrera, Z.; Santana-Rodríguez, J.J. Determination of steroid hormones in fish tissues by microwave-assisted extraction coupled to ultra-high performance liquid chromatography tandem mass spectrometry. Food Chem. 2017, 237, 1012–1020. [Google Scholar] [CrossRef]
- Mokarram, M.; Saber, A.; Sheykhi, V. Effects of heavy metal contamination on river water quality due to release of industrial effluents. J. Clean. Prod. 2020, 277, 123380. [Google Scholar] [CrossRef]
- Long, Z.; Zhu, H.; Bing, H.; Tian, X.; Wang, Z.; Wang, X.; Wu, Y. Contamination, sources and health risk of heavy metals in soil and dust from different functional areas in an industrial city of Panzhihua City, Southwest China. J. Hazard. Mater. 2021, 420, 126638. [Google Scholar] [CrossRef]
- Hoang, H.G.; Lin, C.; Tran, H.T.; Chiang, C.F.; Bui, X.T.; Cheruiyot, N.K.; Shern, C.C.; Lee, C.W. Heavy metal contamination trends in surface water and sediments of a river in a highly-industrialized region. Environ. Technol. Innov. 2020, 20, 101043. [Google Scholar] [CrossRef]
- Ashraf, S.; Rizvi, N.B.; Rasool, A.; Mahmud, T.; Huang, G.G.; Zulfajri, M. Evaluation of heavy metal ions in the groundwater samples from selected automobile workshop areas in northern Pakistan. Groundw. Sustain. Dev. 2020, 11, 100428. [Google Scholar] [CrossRef]
- El-Hak, H.N.G.; El-Din, M.I.S.; Elrayess, R.A. Bioaccumulation of heavy metals and their histopathological impact on Mugil cephalus from the North Eastern Region of Manzala Lake, Egypt. Reg. Stud. Mar. Sci. 2021, 45, 101841. [Google Scholar] [CrossRef]
- Mishra, S.; Bharagava, R.N.; More, N.; Yadav, A.; Zainith, S.; Mani, S.; Chowdhary, P. Heavy Metal Contamination: An Alarming Threat to Environment and Human Health. In Environmental Biotechnology: For Sustainable Future; Springer: Singapore, 2019; pp. 103–125. [Google Scholar] [CrossRef]
- Li, C.; Zhou, K.; Qin, W.; Tian, C.; Qi, M.; Yan, X.; Han, W. A Review on Heavy Metals Contamination in Soil: Effects, Sources, and Remediation Techniques. Soil Sediment Contam. Int. J. 2019, 28, 380–394. [Google Scholar] [CrossRef]
- Devaraj, M.; Sasikumar, Y.; Rajendran, S.; Ponce, L.C. Review—Metal Organic Framework Based Nanomaterials for Electrochemical Sensing of Toxic Heavy Metal Ions: Progress and Their Prospects. J. Electrochem. Soc. 2021, 168, 037513. [Google Scholar] [CrossRef]
- González-González, R.B.; Morales-Murillo, M.B.; Martínez-Prado, M.A.; Melchor-Martínez, E.M.; Ahmed, I.; Bilal, M.; Parra-Saldívar, R.; Iqbal, H.M.N. Carbon dots-based nanomaterials for fluorescent sensing of toxic elements in environmental samples: Strategies for enhanced performance. Chemosphere 2022, 300, 134515. [Google Scholar] [CrossRef]
- Xue, Y.; Wang, Y.; Wang, S.; Yan, M.; Huang, J.; Yang, X. Label-Free and Regenerable Aptasensor for Real-Time Detection of Cadmium(II) by Dual Polarization Interferometry. Anal. Chem. 2020, 92, 10007–10015. [Google Scholar] [CrossRef]
- Stackpoole, S.M.; Shoda, M.E.; Medalie, L.; Stone, W.W. Pesticides in US Rivers: Regional differences in use, occurrence, and environmental toxicity, 2013 to 2017. Sci. Total Environ. 2021, 787, 147147. [Google Scholar] [CrossRef]
- Tan, H.; Zhang, H.; Wu, C.; Wang, C.; Li, Q. Pesticides in surface waters of tropical river basins draining areas with rice–vegetable rotations in Hainan, China: Occurrence, relation to environmental factors, and risk assessment. Environ. Pollut. 2021, 283, 117100. [Google Scholar] [CrossRef] [PubMed]



| Types of Aptasensors | Pollutants Detected | LOD | Range of Detection | Sample Matrix | Reference |
|---|---|---|---|---|---|
| Photoelectrochemical | E. coli O15J_HJ | 200 cfu/mL | 4 × 102 cfu/mL to 4 × 107 cfu/mL | Water | [25] |
| Fluorescence | Salmonella typhimurium | 1 cfu/mL | From 10 to 1010 cfu/mL | Water and food | [26] |
| Colorimetric | Salmonella typhimurium, E. coli and Staphylococcus aureus | ≤104 cfu/mL | NR | Food | [27] |
| Conductometric | SARS-CoV-2 and adenovirus | 1 Pfu/mL for human adenovirus and 1 × 104 copies/mL for SARS-CoV-2 | From 6 pfu/mL to 6 × 104 pfu/mL to adenovirus and 1 × 104 to 1 × 108 copies/mL for SARS-CoV-2 | Water, saliva, and serum | [28] |
| Fluorescence | AMP and KAM | 0.06 ng/L for ampicillin and 0.0150 ng/L for Kanamycin | From 0.5 to 500 ng/L for ampicillin and from 0.5 to 1000 ng/L for Kanamycin | Water | [29] |
| Colorimetric | OTC and KAM | 1 ag/mL | From 10−6 to 105 pg/mL | Food | [30] |
| SERS | DEHP | 8 pM | From 0.008 to 182 nM | Food and water | [31] |
| Colorimetric | DEHP | 1 ng/L | 0.003–10 µg/L | Water | [32] |
| Fluorescence | Bisphenol | 32 nM | 0–1300 µM | Water | [33] |
| SERS | Bisphenol | 0.75 pg/mL | From 0.001 to 100 ng/mL | Water | [34] |
| Electrochemical | Pb2+ | 0.096 µg/L | From 0.1 to 10 µg/L | Water and Soil | [35] |
| Electro-chemiluminescence | Pb2+ | 18.9 pM | 50 pM 387.9 nM | Water | [36] |
| Colorimetric | As3+ | 1.97 ppm for soil samples and 14.44 ppb for aqueous samples | NR | Soil | [37] |
| Fluorescence | As3+ | 5 ng/L | From 10 ng/L to 10 mg/L | Water | [38] |
| Electrochemical | As3+ | 1.4 × 10−7 ng/mL | 3.8 × 10−7−3.0 × 10−4 ng/mL | Water | [39] |
| Fluorescence | Cd2+ | 2.15 nM | From 7.19 nM to 5 µM | Water | [40] |
| Colorimetric | EDI and IBF | 10 nM for edifenphos and 5 nM for Iprobenfos | From 5 to 25 nM for Edifenphos and from 10 to 100 nM for Iprobenfos | Food | [41] |
| Fluorescence | Organophosphorus | 13.4 nM to 23.4 nM | NR | Food | [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores-Contreras, E.A.; González-González, R.B.; González-González, E.; Melchor-Martínez, E.M.; Parra-Saldívar, R.; Iqbal, H.M.N. Detection of Emerging Pollutants Using Aptamer-Based Biosensors: Recent Advances, Challenges, and Outlook. Biosensors 2022, 12, 1078. https://doi.org/10.3390/bios12121078
Flores-Contreras EA, González-González RB, González-González E, Melchor-Martínez EM, Parra-Saldívar R, Iqbal HMN. Detection of Emerging Pollutants Using Aptamer-Based Biosensors: Recent Advances, Challenges, and Outlook. Biosensors. 2022; 12(12):1078. https://doi.org/10.3390/bios12121078
Chicago/Turabian StyleFlores-Contreras, Elda A., Reyna Berenice González-González, Everardo González-González, Elda M. Melchor-Martínez, Roberto Parra-Saldívar, and Hafiz M. N. Iqbal. 2022. "Detection of Emerging Pollutants Using Aptamer-Based Biosensors: Recent Advances, Challenges, and Outlook" Biosensors 12, no. 12: 1078. https://doi.org/10.3390/bios12121078






