Microfluidic Strategies for Extracellular Vesicle Isolation: Towards Clinical Applications
Abstract
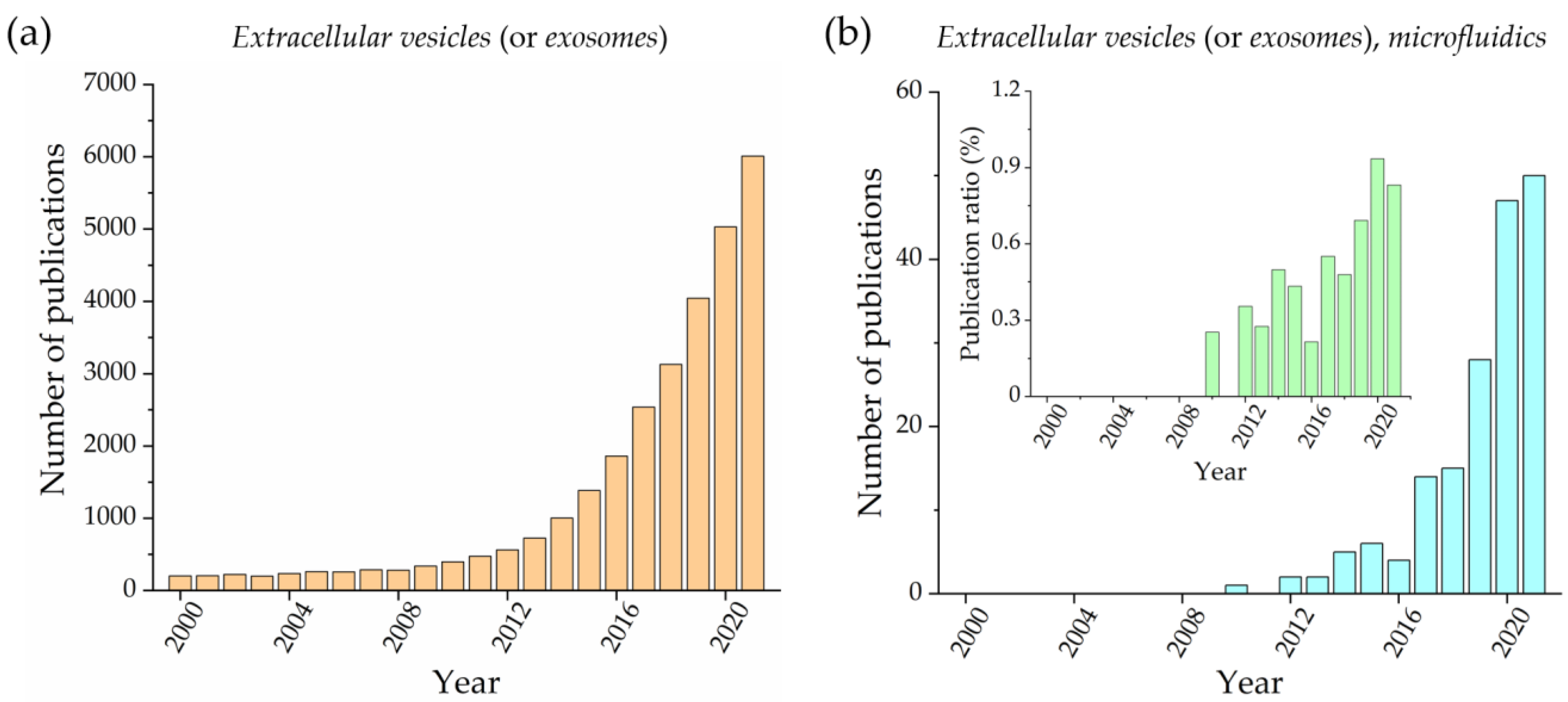
:1. Introduction
2. Conventional EV Isolation Strategies
2.1. Differential Ultracentrifugation and Density Gradient Ultracentrifugation
2.2. Filtration Methods (Ultrafiltration and Size-Exclusion Chromatography)
2.3. Precipitation and Immunoaffinity Methods
2.4. Comments
3. EV Isolation Methods Based on Microfluidic Devices
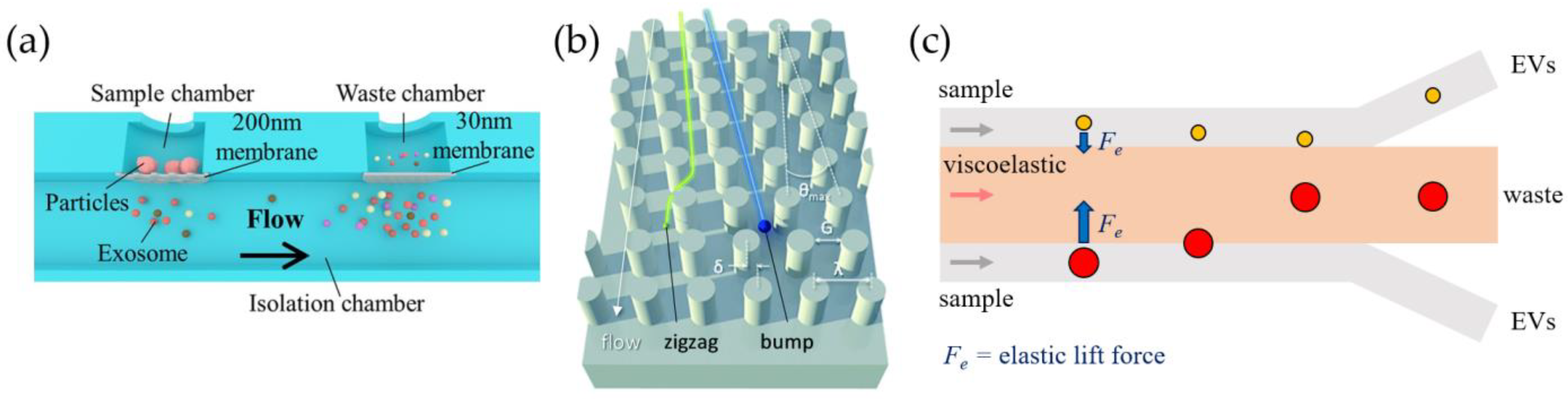
3.1. Physical Methods
3.1.1. Passive Approaches
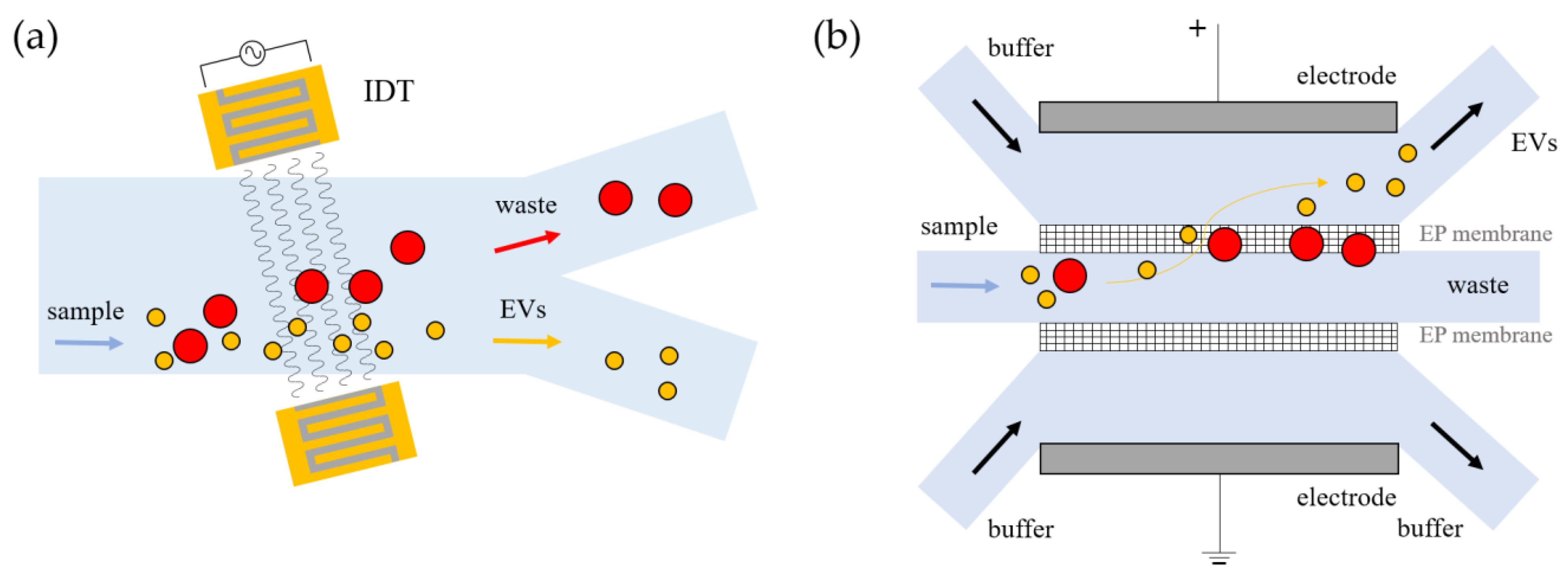
3.1.2. Active Approaches
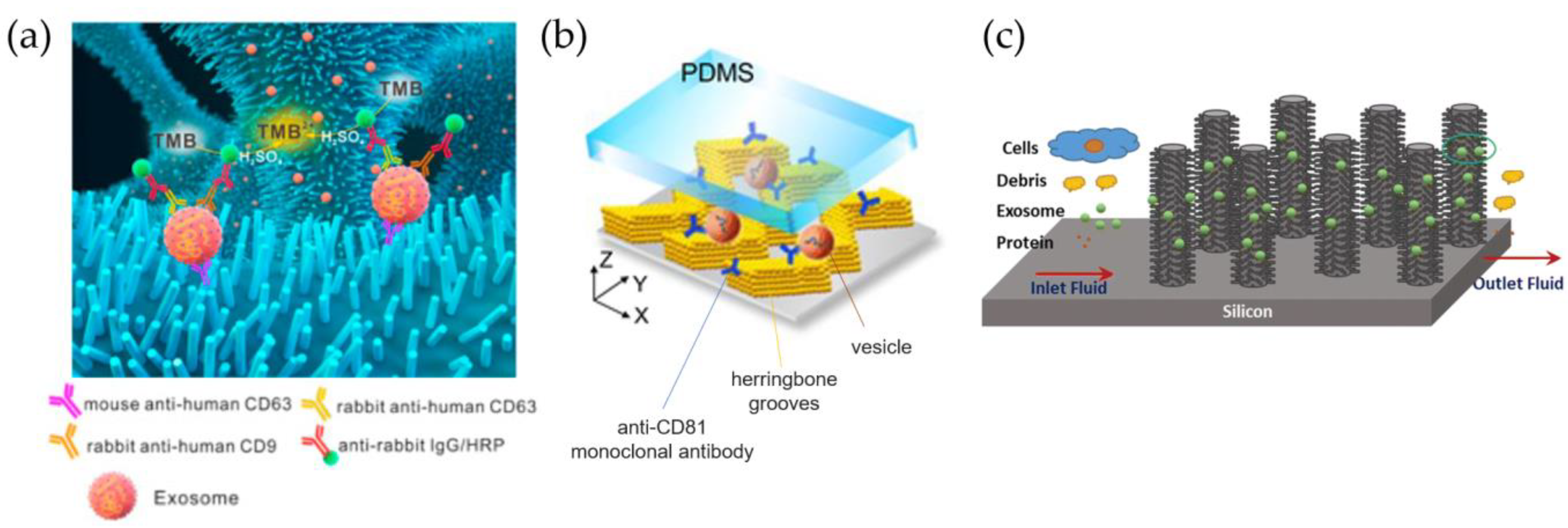
3.2. Chemical Methods
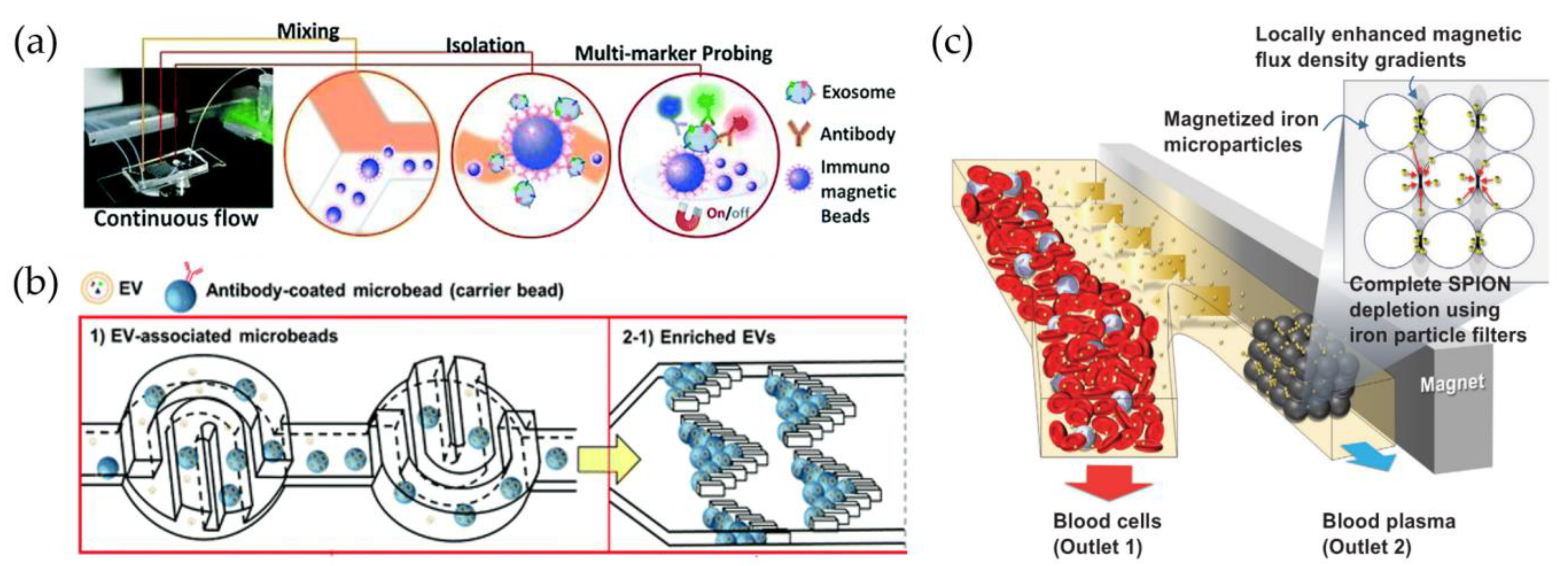
3.2.1. Immunocapture on Fix Support
3.2.2. Immunocapture on Beads and Nanoparticles
4. Discussion
4.1. Physical and Chemical Microfluidic Approaches: Pros and Cons
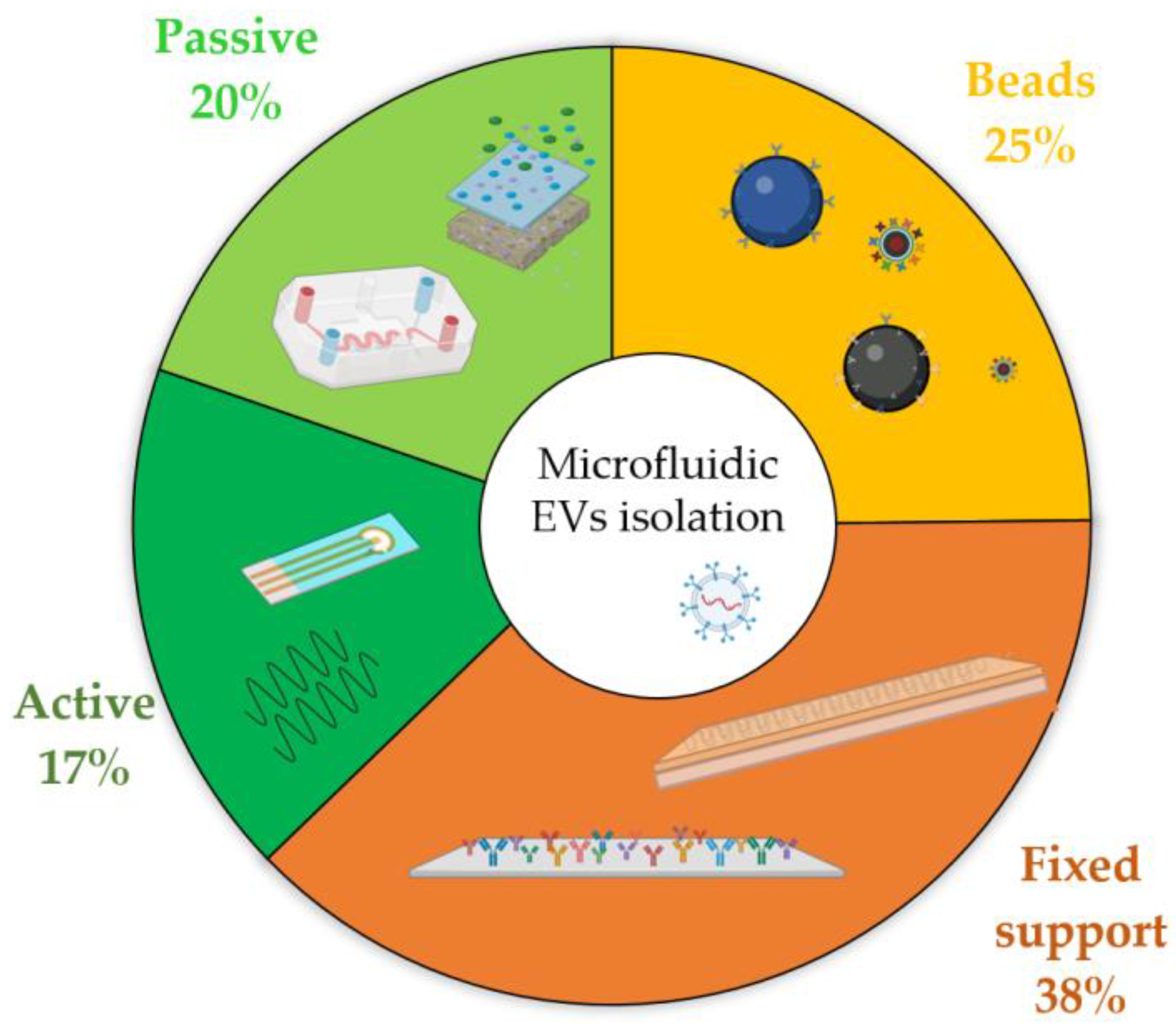
4.2. Microfluidic Isolation Techniques: Which Is the Most Popular
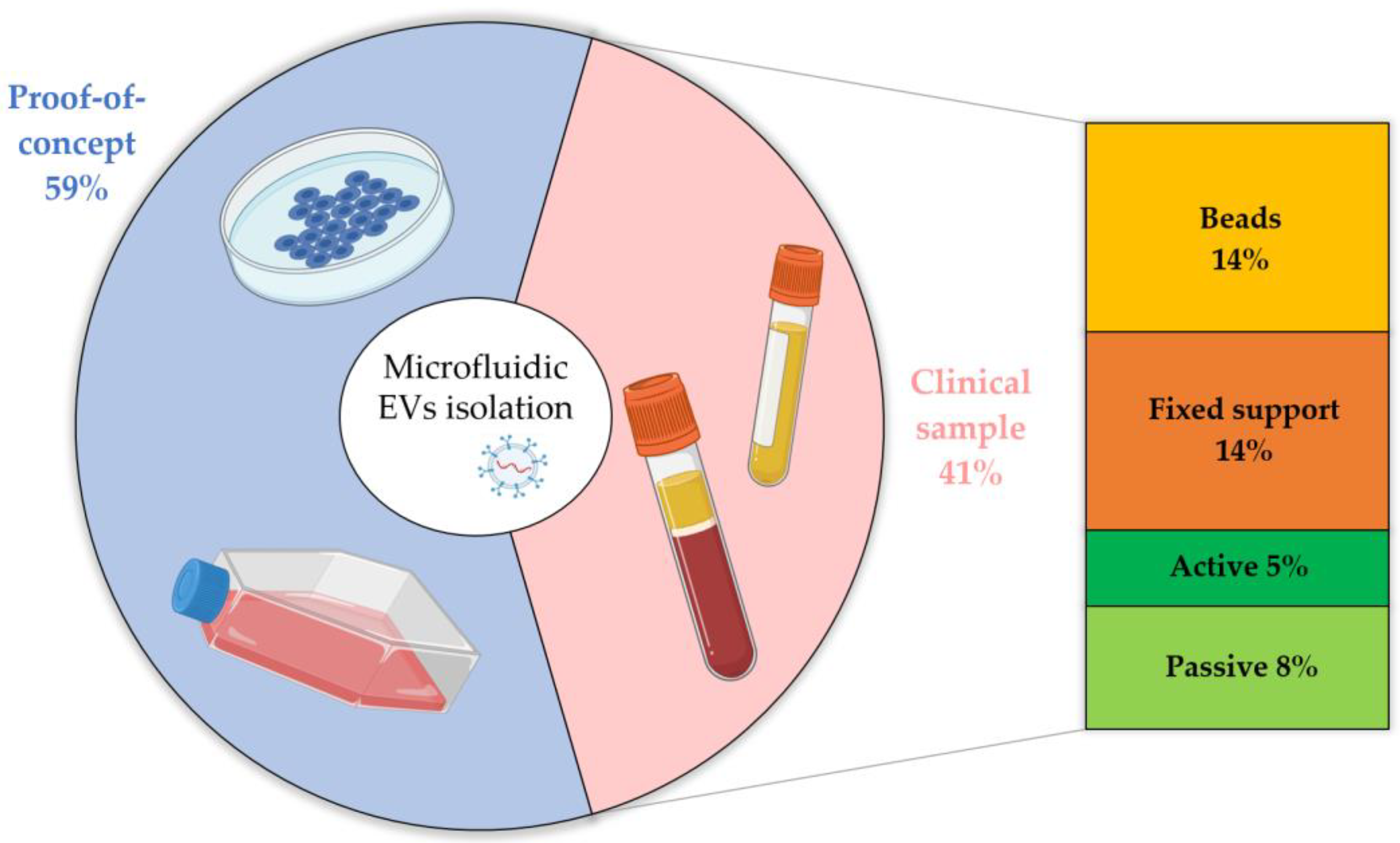
4.3. Are Microfluidic Devices for EV Isolation Ready for Clinical Applications
4.4. Microfluidic Devices for EV Detection and Analysis
5. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ginsburg, G.S.; Phillips, K.A. Precision Medicine: From Science to Value. Health Aff. 2018, 37, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.J.; Ding, L.; Shen, D.; Luo, J.; Suman, V.J.; Wallis, J.W.; Van Tine, B.A.; Hoog, J.; Goiffon, R.J.; Goldstein, T.C.; et al. Whole-Genome Analysis Informs Breast Cancer Response to Aromatase Inhibition. Nature 2012, 486, 353–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loughran, C.; Keeling, C.R. Seeding of Tumour Cells Following Breast Biopsy: A Literature Review. Br. J. Radiol. 2011, 84, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabierèes, C.; Pantel, K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Diaz, L.A.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Stahl, P.D. Extracellular Vesicles: A New Communication Paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510. [Google Scholar] [CrossRef]
- Jia, S.; Zocco, D.; Samuels, M.L.; Chou, M.F.; Chammas, R.; Skog, J.; Zarovni, N.; Momen-Heravi, F.; Kuo, W.P. Emerging Technologies in Extracellular Vesicle-Based Molecular Diagnostics. Expert Rev. Mol. Diagn. 2014, 14, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.; Nickenig, G.; Werner, N. Extracellular Vesicles in Cardiovascular Disease. Circ. Res. 2017, 120, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Turpin, D.; Truchetet, M.E.; Faustin, B.; Augusto, J.F.; Contin-Bordes, C.; Brisson, A.; Blanco, P.; Duffau, P. Role of Extracellular Vesicles in Autoimmune Diseases. Autoimmun. Rev. 2016, 15, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.G.; Gray, E.; Heman-Ackah, S.M.; Mäger, I.; Talbot, K.; El Andaloussi, S.; Wood, M.J.; Turner, M.R. Extracellular Vesicles in Neurodegenerative Disease-Pathogenesis to Biomarkers. Nat. Rev. Neurol. 2016, 12, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Vader, P.; Breakefield, X.O.; Wood, M.J.A. Extracellular Vesicles: Emerging Targets for Cancer Therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Weng, J.; Xiang, X.; Ding, L.; Wong, A.L.A.; Zeng, Q.; Sethi, G.; Wang, L.; Lee, S.C.; Goh, B.C. Extracellular Vesicles, the Cornerstone of next-Generation Cancer Diagnosis? Semin. Cancer Biol. 2021, 74, 105–120. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef] [Green Version]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, Y.; Tang, H.; Zong, N.; Jiang, X. Microfluidics for Biomedical Analysis. Small Methods 2020, 4, 1–30. [Google Scholar] [CrossRef]
- Serrano-Pertierra, E.; Oliveira-Rodríguez, M.; Matos, M.; Gutiérrez, G.; Moyano, A.; Salvador, M.; Rivas, M.; Blanco-López, M.C. Extracellular Vesicles: Current Analytical Techniques for Detection and Quantification. Biomolecules 2020, 10, 824. [Google Scholar] [CrossRef]
- Livshts, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of Exosomes by Differential Centrifugation: Theoretical Analysis of a Commonly Used Protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres Crigna, A.; Fricke, F.; Nitschke, K.; Worst, T.; Erb, U.; Karremann, M.; Buschmann, D.; Elvers-Hornung, S.; Tucher, C.; Schiller, M.; et al. Inter-Laboratory Comparison of Extracellular Vesicle Isolation Based on Ultracentrifugation. Transfus. Med. Hemotherapy 2021, 48, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Kain, S.R. Methods and Protocols; John Wiley & Sons: New York, NY, USA, 2005; Volume 47, ISBN 0471736821. [Google Scholar]
- Grant, R.; Ansa-Addo, E.; Stratton, D.; Antwi-Baffour, S.; Jorfi, S.; Kholia, S.; Krige, L.; Lange, S.; Inal, J. A Filtration-Based Protocol to Isolate Human Plasma Membrane-Derived Vesicles and Exosomes from Blood Plasma. J. Immunol. Methods 2011, 371, 143–151. [Google Scholar] [CrossRef]
- Nordin, J.Z.; Lee, Y.; Vader, P.; Mäger, I.; Johansson, H.J.; Heusermann, W.; Wiklander, O.P.B.; Hällbrink, M.; Seow, Y.; Bultema, J.J.; et al. Ultrafiltration with Size-Exclusion Liquid Chromatography for High Yield Isolation of Extracellular Vesicles Preserving Intact Biophysical and Functional Properties. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 879–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-Step Isolation of Extracellular Vesicles by Size-Exclusion Chromatography. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef]
- Lobb, R.; Möller, A. Size Exclusion Chromatography: A Simple and Reliable Method for Exosome Purification. Methods Mol. Biol. 2017, 1660, 105–110. [Google Scholar] [CrossRef]
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.G.; Vader, P. Higher Functionality of Extracellular Vesicles Isolated Using Size-Exclusion Chromatography Compared to Ultracentrifugation. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2061–2065. [Google Scholar] [CrossRef]
- Monguió-Tortajada, M.; Morón-Font, M.; Gámez-Valero, A.; Carreras-Planella, L.; Borràs, F.E.; Franquesa, M. Extracellular-Vesicle Isolation from Different Biological Fluids by Size-Exclusion Chromatography. Curr. Protoc. Stem Cell Biol. 2019, 49, e82. [Google Scholar] [CrossRef]
- Kaddour, H.; Lyu, Y.; Shouman, N.; Mohan, M.; Okeoma, C.M. Development of Novel High-Resolution Size-Guided Turbidimetry-Enabled Particle Purification Liquid Chromatography (PPLC): Extracellular Vesicles and Membraneless Condensates in Focus. Int. J. Mol. Sci. 2020, 21, 5361. [Google Scholar] [CrossRef]
- Alvarez, F.A.; Kaddour, H.; Lyu, Y.; Preece, C.; Cohen, J.; Baer, L.; Stopeck, A.T.; Thompson, P.; Okeoma, C.M. Blood Plasma Derived Extracellular Vesicles (BEVs): Particle Purification Liquid Chromatography (PPLC) and Proteomic Analysis Reveals BEVs as a Potential Minimally Invasive Tool for Predicting Response to Breast Cancer Treatment. Breast Cancer Res. Treat. 2022, 196, 423–437. [Google Scholar] [CrossRef]
- Niu, Z.; Pang, R.T.K.; Liu, W.; Li, Q.; Cheng, R.; Yeung, W.S.B. Polymer-Based Precipitation Preserves Biological Activities of Extracellular Vesicles from an Endometrial Cell Line. PLoS ONE 2017, 12, e0186534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated Isolation and Quantitative Analysis of Exosome Shuttled Proteins and Nucleic Acids Using Immunocapture Approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Gemoll, T.; Rozanova, S.; Röder, C.; Hartwig, S.; Kalthoff, H.; Lehr, S.; Elsharawy, A.; Habermann, J.K. Protein Profiling of Serum Extracellular Vesicles Reveals Qualitative and Quantitative Differences after Differential Ultracentrifugation and Exoquicktm Isolation. J. Clin. Med. 2020, 9, 1429. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Jakobsen, K.R.; Paulsen, B.S.; Bæk, R.; Varming, K.; Sorensen, B.S.; Jørgensen, M.M. Exosomal Proteins as Potential Diagnostic Markers in Advanced Non-Small Cell Lung Carcinoma. J. Extracell. Vesicles 2015, 4, 1–10. [Google Scholar] [CrossRef]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.G.; Kong, M.Q.; Zhou, S.; Sheng, Y.F.; Wang, P.; Yu, T.; Inci, F.; Kuo, W.P.; Li, L.J.; Demirci, U.; et al. An Integrated Double-Filtration Microfluidic Device for Isolation, Enrichment and Quantification of Urinary Extracellular Vesicles for Detection of Bladder Cancer. Sci. Rep. 2017, 7, 46224. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef]
- Chen, Y.S.; Ma, Y.D.; Chen, C.; Shiesh, S.C.; Lee, G. Bin An Integrated Microfluidic System for On-Chip Enrichment and Quantification of Circulating Extracellular Vesicles from Whole Blood. Lab Chip 2019, 19, 3305–3315. [Google Scholar] [CrossRef]
- Inci, F. Benchmarking a Microfluidic-Based Filtration for Isolating Biological Particles. Langmuir 2022, 38, 1897–1909. [Google Scholar] [CrossRef]
- Riazanski, V.; Mauleon, G.; Lucas, K.; Walker, S.; Zimnicka, A.M.; McGrath, J.L.; Nelson, D.J. Real Time Imaging of Single Extracellular Vesicle PH Regulation in a Microfluidic Cross-Flow Filtration Platform. Commun. Biol. 2022, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Casadei, L.; Choudhury, A.; Sarchet, P.; Mohana Sundaram, P.; Lopez, G.; Braggio, D.; Balakirsky, G.; Pollock, R.; Prakash, S. Cross-Flow Microfiltration for Isolation, Selective Capture and Release of Liposarcoma Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12062. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.K.; Sunkara, V.; Park, J.; Kim, T.H.; Han, J.R.; Kim, C.J.; Choi, H.I.; Kim, Y.K.; Cho, Y.K. Exodisc for Rapid, Size-Selective, and Efficient Isolation and Analysis of Nanoscale Extracellular Vesicles from Biological Samples. ACS Nano 2017, 11, 1360–1370. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, V.; Kim, C.J.; Park, J.; Woo, H.K.; Kim, D.; Ha, H.K.; Kim, M.H.; Son, Y.; Kim, J.R.; Cho, Y.K. Fully Automated, Label-Free Isolation of Extracellular Vesicles from Whole Blood for Cancer Diagnosis and Monitoring. Theranostics 2019, 9, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Wunsch, B.H.; Dogra, N.; Ahsen, M.E.; Lee, K.; Yadav, K.K.; Weil, R.; Pereira, M.A.; Patel, J.V.; Duch, E.A.; et al. Integrated Nanoscale Deterministic Lateral Displacement Arrays for Separation of Extracellular Vesicles from Clinically-Relevant Volumes of Biological Samples. Lab Chip 2018, 18, 3913–3925. [Google Scholar] [CrossRef]
- Segre, G.; Silberg, A. Radial Particle Displacements in Poiseuille Flow of Suspensions. Nature 1961, 189, 209–210. [Google Scholar] [CrossRef]
- Di Carlo, D. Inertial Microfluidics. Lab Chip 2009, 9, 3038–3046. [Google Scholar] [CrossRef]
- Gou, Y.; Jia, Y.; Wang, P.; Sun, C. Progress of Inertial Microfluidics in Principle and Application. Sensors 2018, 18, 1762. [Google Scholar] [CrossRef] [Green Version]
- Yeo, J.C.; Kenry; Zhao, Z.; Zhang, P.; Wang, Z.; Lim, C.T. Label-Free Extraction of Extracellular Vesicles Using Centrifugal Microfluidics. Biomicrofluidics 2018, 12, 024103. [Google Scholar] [CrossRef] [Green Version]
- Kopp, M.R.G.; Linsenmeier, M.; Hettich, B.; Prantl, S.; Stavrakis, S.; Leroux, J.C.; Arosio, P. Microfluidic Shrinking Droplet Concentrator for Analyte Detection and Phase Separation of Protein Solutions. Anal. Chem. 2020, 92, 5803–5812. [Google Scholar] [CrossRef]
- Han, B.H.; Kim, S.; Seo, G.; Heo, Y.; Chung, S.; Kang, J.Y. Isolation of Extracellular Vesicles from Small Volumes of Plasma Using a Microfluidic Aqueous Two-Phase System. Lab Chip 2020, 20, 3552–3559. [Google Scholar] [CrossRef] [PubMed]
- Tay, H.M.; Leong, S.Y.; Xu, X.; Kong, F.; Upadya, M.; Dalan, R.; Tay, C.Y.; Dao, M.; Suresh, S.; Hou, H.W. Direct Isolation of Circulating Extracellular Vesicles from Blood for Vascular Risk Profiling in Type 2 Diabetes Mellitus. Lab Chip 2021, 21, 2511–2523. [Google Scholar] [CrossRef] [PubMed]
- Teoh, B.Y.; Lim, Y.M.; Chong, W.Y.; Subramaniam, M.; Tan, Z.Z.; Misran, M.; Suk, V.R.E.; Lo, K.W.; Lee, P.F. Isolation of Exosome from the Culture Medium of Nasopharyngeal Cancer (NPC) C666-1 Cells Using Inertial Based Microfluidic Channel. Biomed. Microdevices 2022, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Paganini, C.; Hettich, B.; Kopp, M.R.G.; Eördögh, A.; Capasso Palmiero, U.; Adamo, G.; Touzet, N.; Manno, M.; Bongiovanni, A.; Rivera-Fuentes, P.; et al. Rapid Characterization and Quantification of Extracellular Vesicles by Fluorescence-Based Microfluidic Diffusion Sizing. Adv. Healthc. Mater. 2022, 11, 2100021. [Google Scholar] [CrossRef] [PubMed]
- Kuntaegowdanahalli, S.S.; Bhagat, A.A.S.; Kumar, G.; Papautsky, I. Inertial Microfluidics for Continuous Particle Separation in Spiral Microchannels. Lab Chip 2009, 9, 2973–2980. [Google Scholar] [CrossRef] [Green Version]
- Tay, H.M.; Kharel, S.; Dalan, R.; Chen, Z.J.; Tan, K.K.; Boehm, B.O.; Loo, S.C.J.; Hou, H.W. Rapid Purification of Sub-Micrometer Particles for Enhanced Drug Release and Microvesicles Isolation. NPG Asia Mater. 2017, 9, e434. [Google Scholar] [CrossRef] [Green Version]
- McGrath, J.; Jimenez, M.; Bridle, H. Deterministic Lateral Displacement for Particle Separation: A Review. Lab Chip 2014, 14, 4139–4158. [Google Scholar] [CrossRef] [Green Version]
- Inglis, D.W.; Davis, J.A.; Austin, R.H.; Sturm, J.C. Critical Particle Size for Fractionation by Deterministic Lateral Displacement. Lab Chip 2006, 6, 655–658. [Google Scholar] [CrossRef]
- Wunsch, B.H.; Smith, J.T.; Gifford, S.M.; Wang, C.; Brink, M.; Bruce, R.L.; Austin, R.H.; Stolovitzky, G.; Astier, Y. Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids down to 20 Nm. Nat. Nanotechnol. 2016 1111 2016, 11, 936–940. [Google Scholar] [CrossRef]
- Santana, S.M.; Antonyak, M.A.; Cerione, R.A.; Kirby, B.J. Microfluidic Isolation of Cancer-Cell-Derived Microvesicles from Hetergeneous Extracellular Shed Vesicle Populations. Biomed. Microdevices 2014, 16, 869–877. [Google Scholar] [CrossRef]
- Laki, A.J.; Botzheim, L.; Iván, K.; Tamási, V.; Civera, P. Separation of Microvesicles from Serological Samples Using Deterministic Lateral Displacement Effect. Bionanoscience 2015, 5, 48–54. [Google Scholar] [CrossRef]
- Derzsi, L.; Filippi, D.; Mistura, G.; Pierno, M.; Lulli, M.; Sbragaglia, M.; Bernaschi, M.; Garstecki, P. Fluidization and Wall Slip of Soft Glassy Materials by Controlled Surface Roughness. Phys. Rev. E 2017, 95, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leshansky, A.M.; Bransky, A.; Korin, N.; Dinnar, U. Tunable Nonlinear Viscoelastic “Focusing” in a Microfluidic Device. Phys. Rev. Lett. 2007, 98, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.J.; Ober, T.J.; Edd, J.F.; Desai, S.P.; Neal, D.; Bong, K.W.; Doyle, P.S.; McKinley, G.H.; Toner, M. Inertio-Elastic Focusing of Bioparticles in Microchannels at High Throughput. Nat. Commun. 2014, 5, 4120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Xue, C.; Chen, X.; Shan, L.; Tian, Y.; Hu, G. Size-Based Separation of Particles and Cells Utilizing Viscoelastic Effects in Straight Microchannels. Anal. Chem. 2015, 87, 6041–6048. [Google Scholar] [CrossRef] [Green Version]
- Varga, Z.; Fehér, B.; Kitka, D.; Wacha, A.; Bóta, A.; Berényi, S.; Pipich, V.; Fraikin, J.L. Size Measurement of Extracellular Vesicles and Synthetic Liposomes: The Impact of the Hydration Shell and the Protein Corona. Colloids Surfaces B Biointerfaces 2020, 192, 111053. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Z.; Tayebi, M.; Ai, Y. Submicron Particle Focusing and Exosome Sorting by Wavy Microchannel Structures within Viscoelastic Fluids. Anal. Chem. 2019, 91, 4577–4584. [Google Scholar] [CrossRef]
- Nam, J.; Yoon, J.; Jee, H.; Jang, W.S.; Lim, C.S. High-Throughput Separation of Microvesicles from Whole Blood Components Using Viscoelastic Fluid. Adv. Mater. Technol. 2020, 5, 2000612. [Google Scholar] [CrossRef]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhao, J.; Tian, F.; Chang, J.; Zhang, W.; Sun, J. I-DNA- A Nd Aptamer-Mediated Sorting and Analysis of Extracellular Vesicles. J. Am. Chem. Soc. 2019, 141, 3817–3821. [Google Scholar] [CrossRef]
- Asghari, M.; Cao, X.; Mateescu, B.; Van Leeuwen, D.; Aslan, M.K.; Stavrakis, S.; Demello, A.J. Oscillatory Viscoelastic Microfluidics for Efficient Focusing and Separation of Nanoscale Species. ACS Nano 2020, 14, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Fraunhofer, W.; Winter, G. The Use of Asymmetrical Flow Field-Flow Fractionation in Pharmaceutics and Biopharmaceutics. Eur. J. Pharm. Biopharm. 2004, 58, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Wahlund, K.G.; Giddings, J.C. Properties of an Asymmetric Flow Field-Flow, Fractionation Channel Having One Permeable Wall. Anal. Chem. 1987, 59, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Yamada, M.; Nakashima, M.; Seki, M. Pinched Flow Fractionation: Continuous Size Separation of Particles Utilizing a Laminar Flow Profile in a Pinched Microchannel. Anal. Chem. 2004, 76, 5465–5471. [Google Scholar] [CrossRef]
- Shin, S.; Han, D.; Park, M.C.; Mun, J.Y.; Choi, J.; Chun, H.; Kim, S.; Hong, J.W. Separation of Extracellular Nanovesicles and Apoptotic Bodies from Cancer Cell Culture Broth Using Tunable Microfluidic Systems. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Bruus, H. Acoustofluidics 1: Governing Equations in Microfluidics. Lab Chip 2011, 11, 3742–3751. [Google Scholar] [CrossRef] [Green Version]
- Bruus, H. Acoustofluidics 7: The Acoustic Radiation Force on Small Particles. Lab Chip 2012, 12, 1014–1021. [Google Scholar] [CrossRef]
- Bai, Y.; Lu, Y.; Wang, K.; Cheng, Z.; Qu, Y.; Qiu, S.; Zhou, L.; Wu, Z.; Liu, H.; Zhao, J.; et al. Rapid Isolation and Multiplexed Detection of Exosome Tumor Markers Via Queued Beads Combined with Quantum Dots in a Microarray. Nano-Micro Lett. 2019, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sehgal, P.; Kirby, B.J. Separation of 300 and 100 Nm Particles in Fabry-Perot Acoustofluidic Resonators. Anal. Chem. 2017, 89, 12192–12200. [Google Scholar] [CrossRef]
- Leibacher, I.; Reichert, P.; Dual, J. Microfluidic Droplet Handling by Bulk Acoustic Wave (BAW) Acoustophoresis. Lab Chip 2015, 15, 2896–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wu, M.; Lin, Y.; Xu, J. Acoustic Microfluidic Separation Techniques and Bioapplications: A Review. Micromachines 2020, 11, 921. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Li, P.; Lin, S.C.S.; Stratton, Z.S.; Nama, N.; Guo, F.; Slotcavage, D.; Mao, X.; Shi, J.; Costanzo, F.; et al. Surface Acoustic Wave Microfluidics. Lab Chip 2013, 13, 3626–3649. [Google Scholar] [CrossRef]
- Wu, M.; Ozcelik, A.; Rufo, J.; Wang, Z.; Fang, R.; Jun Huang, T. Acoustofluidic Separation of Cells and Particles. Microsystems Nanoeng. 2019, 5, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Shao, H.; Weissleder, R.; Lee, H. Acoustic Purification of Extracellular Microvesicles. ACS Nano 2015, 9, 2321–2327. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Chen, C.; Mao, Z.; Bachman, H.; Becker, R.; Rufo, J.; Wang, Z.; Zhang, P.; Mai, J.; Yang, S.; et al. Acoustofluidic Centrifuge for Nanoparticle Enrichment and Separation. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Wang, Z.; Li, F.; Rufo, J.; Chen, C.; Yang, S.; Li, L.; Zhang, J.; Cheng, J.; Kim, Y.; Wu, M.; et al. Acoustofluidic Salivary Exosome Isolation: A Liquid Biopsy Compatible Approach for Human Papillomavirus–Associated Oropharyngeal Cancer Detection. J. Mol. Diagnostics 2020, 22, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Habibi, R.; He, V.; Ghavamian, S.; De Marco, A.; Lee, T.H.; Aguilar, M.I.; Zhu, D.; Lim, R.; Neild, A. Exosome Trapping and Enrichment Using a Sound Wave Activated Nano-Sieve (SWANS). Lab Chip 2020, 20, 3633–3643. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of Exosomes from Whole Blood by Integrating Acoustics and Microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [Green Version]
- Ku, A.; Lim, H.C.; Evander, M.; Lilja, H.; Laurell, T.; Scheding, S.; Ceder, Y. Acoustic Enrichment of Extracellular Vesicles from Biological Fluids. Anal. Chem. 2018, 90, 8011–8019. [Google Scholar] [CrossRef]
- Ku, A.; Fredsøe, J.; Sørensen, K.D.; Borre, M.; Evander, M.; Laurell, T.; Lilja, H.; Ceder, Y. High-Throughput and Automated Acoustic Trapping of Extracellular Vesicles to Identify MicroRNAs With Diagnostic Potential for Prostate Cancer. Front. Oncol. 2021, 11, 386. [Google Scholar] [CrossRef] [PubMed]
- Ku, A.; Ravi, N.; Yang, M.; Evander, M.; Laurell, T.; Lilja, H.; Ceder, Y. A Urinary Extracellular Vesicle MicroRNA Biomarker Discovery Pipeline; from Automated Extracellular Vesicle Enrichment by Acoustic Trapping to MicroRNA Sequencing. PLoS ONE 2019, 14, e0217507. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Pei, Z.; Liu, P.; Bachman, H.; Downing Naquin, T.; Zhang, P.; Zhang, J.; Shen, L.; Yang, S.; Yang, K.; et al. Acoustofluidics-Assisted Fluorescence-SERS Bimodal Biosensors. Small 2020, 16, 2005179. [Google Scholar] [CrossRef]
- Hao, N.; Liu, P.; Bachman, H.; Pei, Z.; Zhang, P.; Rufo, J.; Wang, Z.; Zhao, S.; Huang, T.J. Acoustofluidics-Assisted Engineering of Multifunctional Three-Dimensional Zinc Oxide Nanoarrays. ACS Nano 2020, 14, 6150–6163. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, C.; Jin, D.; Yu, Y.; Yang, F.; Zhang, Y.; Yao, Q.; Zhang, G.J. AuNP-Amplified Surface Acoustic Wave Sensor for the Quantification of Exosomes. ACS Sens. 2020, 5, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Jubery, T.Z.; Srivastava, S.K.; Dutta, P. Dielectrophoretic Separation of Bioparticles in Microdevices: A Review. Electrophoresis 2014, 35, 691–713. [Google Scholar] [CrossRef]
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C.; et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 2017, 11, 6641–6651. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, L.; Ye, Y.; Li, Y.; Luan, X.; Liu, J.; Cheng, J.; Zhao, Y.; Li, M.; Huang, C. Microsphere Mediated Exosome Isolation and Ultra-Sensitive Detection on a Dielectrophoresis Integrated Microfluidic Device. Analyst 2021, 146, 5962–5972. [Google Scholar] [CrossRef]
- Chen, Y.S.; Lai, C.P.K.; Chen, C.; Lee, G. Bin Isolation and Recovery of Extracellular Vesicles Using Optically-Induced Dielectrophoresis on an Integrated Microfluidic Platform. Lab Chip 2021, 21, 1475–1483. [Google Scholar] [CrossRef]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.J.; Gho, Y.S.; Park, J. Microfluidic Filtration System to Isolate Extracellular Vesicles from Blood. Lab Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef]
- Akagi, T.; Kato, K.; Kobayashi, M.; Kosaka, N.; Ochiya, T.; Ichiki, T. On-Chip Immunoelectrophoresis of Extracellular Vesicles Released from Human Breast Cancer Cells. PLoS ONE 2015, 10, e0123603. [Google Scholar] [CrossRef] [PubMed]
- Marczak, S.; Richards, K.; Ramshani, Z.; Smith, E.; Senapati, S.; Hill, R.; Go, D.B.; Chang, H.C. Simultaneous Isolation and Preconcentration of Exosomes by Ion Concentration Polarization. Electrophoresis 2018, 39, 2029–2038. [Google Scholar] [CrossRef] [PubMed]
- Hadady, H.; Karamali, F.; Ejeian, F.; Haghjooy Javanmard, S.; Rafiee, L.; Nasr Esfahani, M.H. AC Electrokinetic Isolation and Detection of Extracellular Vesicles from Dental Pulp Stem Cells: Theoretical Simulation Incorporating Fluid Mechanics. Electrophoresis 2021, 42, 2018–2026. [Google Scholar] [CrossRef] [PubMed]
- Cheung, L.S.; Sahloul, S.; Orozaliev, A.; Song, Y.A. Rapid Detection and Trapping of Extracellular Vesicles by Electrokinetic Concentration for Liquid Biopsy on Chip. Micromachines 2018, 9, 306. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.; Jo, W.; Heo, Y.; Kang, J.Y.; Kwak, R.; Park, J. Isolation of Extracellular Vesicle from Blood Plasma Using Electrophoretic Migration through Porous Membrane. Sens. Actuators B Chem. 2016, 233, 289–297. [Google Scholar] [CrossRef]
- Tayebi, M.; Yang, D.; Collins, D.J.; Ai, Y. Deterministic Sorting of Submicrometer Particles and Extracellular Vesicles Using a Combined Electric and Acoustic Field. Nano Lett. 2021, 21, 6835–6842. [Google Scholar] [CrossRef]
- Zhang, Y.; Tong, X.; Yang, L.; Yin, R.; Li, Y.; Zeng, D.; Wang, X.; Deng, K. A Herringbone Mixer Based Microfluidic Device HBEXO-Chip for Purifying Tumor-Derived Exosomes and Establishing MiRNA Signature in Pancreatic Cancer. Sens. Actuators B Chem. 2021, 332, 129511. [Google Scholar] [CrossRef]
- Chen, Z.; Cheng, S.B.; Cao, P.; Qiu, Q.F.; Chen, Y.; Xie, M.; Xu, Y.; Huang, W.H. Detection of Exosomes by ZnO Nanowires Coated Three-Dimensional Scaffold Chip Device. Biosens. Bioelectron. 2018, 122, 211–216. [Google Scholar] [CrossRef]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic Device (ExoChip) for on-Chip Isolation, Quantification and Characterization of Circulating Exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Hisey, C.L.; Dorayappan, K.D.P.; Cohn, D.E.; Selvendiran, K.; Hansford, D.J. Microfluidic Affinity Separation Chip for Selective Capture and Release of Label-Free Ovarian Cancer Exosomes. Lab Chip 2018, 18, 3144–3153. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, H.J.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.J.; Liu, X. Ciliated Micropillars for the Microfluidic-Based Isolation of Nanoscale Lipid Vesicles. Lab Chip 2013, 13, 2879–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, R.; Zhu, G.; Wang, Y.; Wu, S.; Li, S.; Zhang, D.; Bu, Y.; Bhave, G.; Han, R.; Liu, X. Microfluidic Device for the Analysis of MDR Cancerous Cell-Derived Exosomes’ Response to Nanotherapy. Biomed. Microdevices 2019, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kamyabi, N.; Abbasgholizadeh, R.; Maitra, A.; Ardekani, A.; Biswal, S.L.; Grande-Allen, K.J. Isolation and Mutational Assessment of Pancreatic Cancer Extracellular Vesicles Using a Microfluidic Platform. Biomed. Microdevices 2020, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sooriyaarachchi, D.; Maharubin, S.; Tan, G.Z. ZnO Nanowire-Anchored Microfluidic Device With Herringbone Structure Fabricated by Maskless Photolithography. Biomed. Eng. Comput. Biol. 2020, 11, 1179597220941431. [Google Scholar] [CrossRef]
- Yang, L.; Tong, X.; Zhang, Y.; Li, Y.; Liu, J.; Yin, R.; Zeng, D.; Yuan, Y.; Deng, K. Tim4-Functionalized HBEV-Chip by Isolating Plasma-Derived Phosphatidylserine-Positive Small Extracellular Vesicles for Pan-Cancer Screening. Adv. Mater. Technol. 2022, 7, 2101115. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Xue, Y.; Qiao, L.; Yu, G.; Liu, Y.; Yu, S. Ultrasensitive Analysis of Exosomes Using a 3D Self-Assembled Nanostructured SiO2Microfluidic Chip. ACS Appl. Mater. Interfaces 2022, 14, 14693–14702. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Bian, F.; Guo, J.; Zhao, Y.; Chen, H.X.; Bian, F.K.; Guo, J.H.; Zhao, Y.J. Aptamer-Functionalized Barcodes in Herringbone Microfluidics for Multiple Detection of Exosomes. Small Methods 2022, 6, 2200236. [Google Scholar] [CrossRef]
- Zhang, P.; He, M.; Zeng, Y. Ultrasensitive Microfluidic Analysis of Circulating Exosomes Using a Nanostructured Graphene Oxide/Polydopamine Coating. Lab Chip 2016, 16, 3033–3042. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.-T.; Hadlock, T.; Lo, T.-W.; Purcell, E.; Mutukuri, A.; Fouladdel, S.; De Silva Raguera, M.; Fairbairn, H.; Murlidhar, V.; Durham, A.; et al. Dual-Isolation and Profiling of Circulating Tumor Cells and Cancer Exosomes from Blood Samples with Melanoma Using Immunoaffinity-Based Microfluidic Interfaces. Adv. Sci. 2020, 7, 2001581. [Google Scholar] [CrossRef]
- Xiong, Y.; Kang, H.; Zhou, H.; Ma, L.; Xu, X. Recent Progress on Microfluidic Devices with Incorporated 1D Nanostructures for Enhanced Extracellular Vesicle (EV) Separation. Bio-Design Manuf. 2022, 5, 607–616. [Google Scholar] [CrossRef]
- Chen, C.; Skog, J.; Hsu, C.H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic Isolation and Transcriptome Analysis of Serum Microvesicles. Lab Chip 2010, 10, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Zhou, X.; He, M.; Shang, Y.; Tetlow, A.L.; Godwin, A.K.; Zeng, Y. Ultrasensitive Detection of Circulating Exosomes with a 3D-Nanopatterned Microfluidic Chip. Nat. Biomed. Eng. 2019, 3, 438–451. [Google Scholar] [CrossRef]
- Gwak, H.; Park, S.; Kim, J.; Lee, J.D.; Kim, I.S.; Kim, S.I.; Hyun, K.A.; Jung, H. Il Microfluidic Chip for Rapid and Selective Isolation of Tumor-Derived Extracellular Vesicles for Early Diagnosis and Metastatic Risk Evaluation of Breast Cancer. Biosens. Bioelectron. 2021, 192, 113495. [Google Scholar] [CrossRef]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated Immunoisolation and Protein Analysis of Circulating Exosomes Using Microfluidic Technology. Lab Chip 2014, 14, 3773–3780. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection towards Blood-Based Ovarian Cancer Diagnosis. Lab Chip 2016, 16, 489–496. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Ye, L.; Jian, X.; Yang, D.; Zhang, H.; Tong, Z.; Wu, Z.; Shi, N.; Han, Y.; Mao, H. Integrated Microfluidic System for Isolating Exosome and Analyzing Protein Marker PD-L1. Biosens. Bioelectron. 2022, 204, 113879. [Google Scholar] [CrossRef]
- Tian, Q.; He, C.; Liu, G.; Zhao, Y.; Hui, L.; Mu, Y.; Tang, R.; Luo, Y.; Zheng, S.; Wang, B. Nanoparticle Counting by Microscopic Digital Detection: Selective Quantitative Analysis of Exosomes via Surface-Anchored Nucleic Acid Amplification. Anal. Chem. 2018, 90, 6556–6562. [Google Scholar] [CrossRef]
- Cheng, H.L.; Fu, C.Y.; Kuo, W.C.; Chen, Y.W.; Chen, Y.S.; Lee, Y.M.; Li, K.H.; Chen, C.; Ma, H.P.; Huang, P.C.; et al. Detecting MiRNA Biomarkers from Extracellular Vesicles for Cardiovascular Disease with a Microfluidic System. Lab Chip 2018, 18, 2917–2925. [Google Scholar] [CrossRef]
- Reátegui, E.; Van Der Vos, K.E.; Lai, C.P.; Zeinali, M.; Atai, N.A.; Aldikacti, B.; Floyd, F.P.; Khankhel, A.; Thapar, V.; Hochberg, F.H.; et al. Engineered Nanointerfaces for Microfluidic Isolation and Molecular Profiling of Tumor-Specific Extracellular Vesicles. Nat. Commun. 2018, 9, 175. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Liao, C.; Zuo, P.; Liu, Z.; Ye, B.C. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-Based Isolation of Melanoma Cell-Derived Exosomes from Plasma of Patients with Melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef]
- Chen, W.; Li, H.; Su, W.; Qin, J. Microfluidic Device for On-Chip Isolation and Detection of Circulating Exosomes in Blood of Breast Cancer Patients. Biomicrofluidics 2019, 13, 054113. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Q.; Shi, H.; Tang, K.; Qiao, L.; Yu, G.; Ding, C.; Yu, S. Microfluidic Raman Biochip Detection of Exosomes: A Promising Tool for Prostate Cancer Diagnosis. Lab Chip 2020, 20, 4632–4637. [Google Scholar] [CrossRef]
- Sung, C.Y.; Huang, C.C.; Chen, Y.S.; Hsu, K.F.; Lee, G. Bin Isolation and Quantification of Extracellular Vesicle-Encapsulated MicroRNA on an Integrated Microfluidic Platform. Lab Chip 2021, 21, 4660–4671. [Google Scholar] [CrossRef]
- Dueck, J. The Sedimentation Velocity of a Particle in a Wide Range of Reynolds Numbers in the Application to the Analysis of the Separation Curve. Adv. Powder Technol. 2013, 24, 150–153. [Google Scholar] [CrossRef]
- Gwak, H.; Park, S.; Yu, H.; Hyun, K.A.; Jung, H. Il A Modular Microfluidic Platform for Serial Enrichment and Harvest of Pure Extracellular Vesicles. Analyst 2022, 147, 1117–1127. [Google Scholar] [CrossRef]
- Kwon, S.; Oh, J.; Seok Lee, M.; Um, E.; Jeong, J.; Kang, J.H.; Kwon, S.; Oh, J.; Lee, M.S.; Kang, J.H.; et al. Enhanced Diamagnetic Repulsion of Blood Cells Enables Versatile Plasma Separation for Biomarker Analysis in Blood. Small 2021, 17, 2100797. [Google Scholar] [CrossRef]
- Son, K.J.; Rahimian, A.; Shin, D.S.; Siltanen, C.; Patel, T.; Revzin, A. Microfluidic Compartments with Sensing Microbeads for Dynamic Monitoring of Cytokine and Exosome Release from Single Cells. Analyst 2016, 141, 679–688. [Google Scholar] [CrossRef]
- Tayebi, M.; Zhou, Y.; Tripathi, P.; Chandramohanadas, R.; Ai, Y. Exosome Purification and Analysis Using a Facile Microfluidic Hydrodynamic Trapping Device. Anal. Chem. 2020, 92, 10733–10742. [Google Scholar] [CrossRef]
- Dudani, J.S.; Gossett, D.R.; Tse, H.T.K.; Lamm, R.J.; Kulkarni, R.P.; Carlo, D. Di Rapid Inertial Solution Exchange for Enrichment and Flow Cytometric Detection of Microvesicles. Biomicrofluidics 2015, 9, 014112. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef]
- Hong, S.L.; Wan, Y.T.; Tang, M.; Pang, D.W.; Zhang, Z.L. Multifunctional Screening Platform for the Highly Efficient Discovery of Aptamers with High Affinity and Specificity. Anal. Chem. 2017, 89, 6535–6542. [Google Scholar] [CrossRef]
- Ko, J.; Bhagwat, N.; Yee, S.S.; Ortiz, N.; Sahmoud, A.; Black, T.; Aiello, N.M.; McKenzie, L.; O’Hara, M.; Redlinger, C.; et al. Combining Machine Learning and Nanofluidic Technology to Diagnose Pancreatic Cancer Using Exosomes. ACS Nano 2017, 11, 11182–11193. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, W.; Cheng, R.; Logun, M.; Zayas-Viera, M.D.M.; Karumbaiah, L.; Mao, L. Label-Free Ferrohydrodynamic Separation of Exosome-like Nanoparticles. Lab Chip 2020, 20, 3187–3201. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Sebastián, V.; Sesé, J.; Pazo-Cid, R.; Mendoza, G.; Arruebo, M.; Martín-Duque, P.; Santamaría, J. Isolation of Exosomes from Whole Blood by a New Microfluidic Device: Proof of Concept Application in the Diagnosis and Monitoring of Pancreatic Cancer. J. Nanobiotechnol. 2020, 18, 150. [Google Scholar] [CrossRef]
- Yu, Z.; Lin, S.; Xia, F.; Liu, Y.; Zhang, D.; Wang, F.; Wang, Y.; Li, Q.; Niu, J.; Cao, C.; et al. ExoSD Chips for High-Purity Immunomagnetic Separation and High-Sensitivity Detection of Gastric Cancer Cell-Derived Exosomes. Biosens. Bioelectron. 2021, 194, 113594. [Google Scholar] [CrossRef]
- Yang, Y.; Kannisto, E.; Patnaik, S.K.; Reid, M.E.; Li, L.; Wu, Y. Ultrafast Detection of Exosomal RNAs via Cationic Lipoplex Nanoparticles in a Micromixer Biochip for Cancer Diagnosis. ACS Appl. Nano Mater. 2021, 4, 2806–2819. [Google Scholar] [CrossRef]
- Vader, P.; Mol, E.A.; Pasterkamp, G.; Schiffelers, R.M. Extracellular Vesicles for Drug Delivery. Adv. Drug Deliv. Rev. 2016, 106, 148–156. [Google Scholar] [CrossRef]
- Mason, H.G.; Bush, J.; Agrawal, N.; Hakami, R.M.; Veneziano, R. A Microfluidic Platform to Monitor Real-Time Effects of Extracellular Vesicle Exchange between Co-Cultured Cells across Selectively Permeable Barriers. Int. J. Mol. Sci. 2022, 23, 3534. [Google Scholar] [CrossRef]
- Han, Z.; Peng, C.; Yi, J.; Zhang, D.; Xiang, X.; Peng, X.; Su, B.; Liu, B.; Shen, Y.; Qiao, L. Highly Efficient Exosome Purification from Human Plasma by Tangential Flow Filtration Based Microfluidic Chip. Sensors Actuators B Chem. 2021, 333, 129563. [Google Scholar] [CrossRef]
- Gustafson, K.T.; Huynh, K.T.; Heineck, D.; Bueno, J.; Modestino, A.; Kim, S.; Gower, A.; Armstrong, R.; Schutt, C.E.; Ibsen, S.D. Automated Fluorescence Quantification of Extracellular Vesicles Collected from Blood Plasma Using Dielectrophoresis. Lab Chip 2021, 21, 1318–1332. [Google Scholar] [CrossRef]
- Mogi, K.; Hayashida, K.; Yamamoto, T. Damage-Less Handling of Exosomes Using an Ion-Depletion Zone in a Microchannel. Anal. Sci. 2018, 34, 875–880. [Google Scholar] [CrossRef] [Green Version]
- Vaidyanathan, R.; Naghibosadat, M.; Rauf, S.; Korbie, D.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Detecting Exosomes Specifically: A Multiplexed Device Based on Alternating Current Electrohydrodynamic Induced Nanoshearing. Anal. Chem. 2014, 86, 11125–11132. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.-T.; Purcell, E.; Palacios-Rolston, C.; Lo, T.-W.; Ramnath, N.; Jolly, S.; Nagrath, S.; Kang, Y.; Purcell, E.; Palacios-Rolston, C.; et al. Isolation and Profiling of Circulating Tumor-Associated Exosomes Using Extracellular Vesicular Lipid–Protein Binding Affinity Based Microfluidic Device. Small 2019, 15, 1903600. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, Y.; Qian, X. Target-Specific Exosome Isolation through Aptamer-Based Microfluidics. Biosensors 2022, 12, 257. [Google Scholar] [CrossRef]
- Kang, Y.T.; Purcell, E.; Hadlock, T.; Lo, T.W.; Mutukuri, A.; Jolly, S.; Nagrath, S. Multiplex Isolation and Profiling of Extracellular Vesicles Using a Microfluidic DICE Device. Analyst 2019, 144, 5785–5793. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, K.; Cui, J.; Liu, H.; Bu, X.; Ma, H.; Wang, W.; Gong, H.; Lausted, C.; Hood, L.; et al. Label-Free Quantitative Detection of Tumor-Derived Exosomes through Surface Plasmon Resonance Imaging. Anal. Chem. 2014, 86, 8857–8864. [Google Scholar] [CrossRef] [Green Version]
- Yasui, T.; Yanagida, T.; Ito, S.; Konakade, Y.; Takeshita, D.; Naganawa, T.; Nagashima, K.; Shimada, T.; Kaji, N.; Nakamura, Y.; et al. Unveiling Massive Numbers of Cancer-Related Urinary-MicroRNA Candidates via Nanowires. Sci. Adv. 2017, 3. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Li, W.; Zhang, L.; Ban, L.; Chen, P.; Du, W.; Feng, X.; Liu, B.F. Chemically Edited Exosomes with Dual Ligand Purified by Microfluidic Device for Active Targeted Drug Delivery to Tumor Cells. ACS Appl. Mater. Interfaces 2017, 9, 27441–27452. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, W.; Kimber, M.; Lu, M.; Dong, L. Rapid Differentiation of Host and Parasitic Exosome Vesicles Using Microfluidic Photonic Crystal Biosensor. ACS Sensors 2018, 3, 1616–1621. [Google Scholar] [CrossRef]
- Algarni, A.; Greenman, J.; Madden, L.A. Procoagulant Tumor Microvesicles Attach to Endothelial Cells on Biochips under Microfluidic Flow. Biomicrofluidics 2019, 13, 064124. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, X.; Zeng, Y. Multiplexed Immunophenotyping of Circulating Exosomes on Nano-Engineered ExoProfile Chip towards Early Diagnosis of Cancer. Chem. Sci. 2019, 10, 5495–5504. [Google Scholar] [CrossRef] [Green Version]
- Cavallaro, S.; Horak, J.; HÅÅg, P.; Gupta, D.; Stiller, C.; Sahu, S.S.; Görgens, A.; Gatty, H.K.; Viktorsson, K.; El Andaloussi, S.; et al. Label-Free Surface Protein Profiling of Extracellular Vesicles by an Electrokinetic Sensor. ACS Sens. 2019, 4, 1399–1408. [Google Scholar] [CrossRef]
- Lv, X.; Geng, Z.; Su, Y.; Fan, Z.; Wang, S.; Fang, W.; Chen, H. Label-Free Exosome Detection Based on a Low-Cost Plasmonic Biosensor Array Integrated with Microfluidics. Langmuir 2019, 35, 9816–9824. [Google Scholar] [CrossRef]
- Wijerathne, H.; Witek, M.A.; Jackson, J.M.; Brown, V.; Hupert, M.L.; Herrera, K.; Kramer, C.; Davidow, A.E.; Li, Y.; Baird, A.E.; et al. Affinity Enrichment of Extracellular Vesicles from Plasma Reveals MRNA Changes Associated with Acute Ischemic Stroke. Commun. Biol. 2020, 3, 613. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, T.; Han, G.; Wu, Y.; Hua, X.; Su, J.; Jin, W.; Mou, Y.; Mou, X.; Li, Q.; et al. Accurate Cancer Diagnosis and Stage Monitoring Enabled by Comprehensive Profiling of Different Types of Exosomal Biomarkers: Surface Proteins and MiRNAs. Small 2020, 16, 2004492. [Google Scholar] [CrossRef]
- Han, S.; Xu, Y.; Sun, J.; Liu, Y.; Zhao, Y.; Tao, W.; Chai, R. Isolation and Analysis of Extracellular Vesicles in a Morpho Butterfly Wing-Integrated Microvortex Biochip. Biosens. Bioelectron. 2020, 154, 112073. [Google Scholar] [CrossRef]
- Yang, Q.; Cheng, L.; Hu, L.; Lou, D.; Zhang, T.; Li, J.; Zhu, Q.; Liu, F. An Integrative Microfluidic Device for Isolation and Ultrasensitive Detection of Lung Cancer-Specific Exosomes from Patient Urine. Biosens. Bioelectron. 2020, 163, 112290. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, T.; Zhang, F.; Tang, D.; Li, D.; Cao, J.; Wei, W.; Wu, Y.; Liu, S. Integrated Microfluidic Device for Accurate Extracellular Vesicle Quantification and Protein Markers Analysis Directly from Human Whole Blood. Anal. Chem. 2020, 92, 1574–1581. [Google Scholar] [CrossRef]
- Kashefi-Kheyrabadi, L.; Kim, J.; Chakravarty, S.; Park, S.; Gwak, H.; Kim, S.I.; Mohammadniaei, M.; Lee, M.H.; Hyun, K.A.; Jung, H. Il Detachable Microfluidic Device Implemented with Electrochemical Aptasensor (DeMEA) for Sequential Analysis of Cancerous Exosomes. Biosens. Bioelectron. 2020, 169, 112622. [Google Scholar] [CrossRef]
- Yu, Q.; Zhao, Q.; Wang, S.; Zhao, S.; Zhang, S.; Yin, Y.; Dong, Y. Development of a Lateral Flow Aptamer Assay Strip for Facile Identification of Theranostic Exosomes Isolated from Human Lung Carcinoma Cells. Anal. Biochem. 2020, 594, 113591. [Google Scholar] [CrossRef]
- Sun, N.; Lee, Y.T.; Zhang, R.Y.; Kao, R.; Teng, P.C.; Yang, Y.; Yang, P.; Wang, J.J.; Smalley, M.; Chen, P.J.; et al. Purification of HCC-Specific Extracellular Vesicles on Nanosubstrates for Early HCC Detection by Digital Scoring. Nat. Commun. 2020, 11, 4489. [Google Scholar] [CrossRef]
- Nikoloff, J.M.; Saucedo-Espinosa, M.A.; Kling, A.; Dittrich, P.S. Identifying Extracellular Vesicle Populations from Single Cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2106630118. [Google Scholar] [CrossRef]
- Chiodi, E.; Daaboul, G.G.; Marn, A.M.; Ünlü, M.S. Multiplexed Affinity Measurements of Extracellular Vesicles Binding Kinetics. Sensors 2021, 21, 2634. [Google Scholar] [CrossRef]
- Suwatthanarak, T.; Thiodorus, I.A.; Tanaka, M.; Shimada, T.; Takeshita, D.; Yasui, T.; Baba, Y.; Okochi, M. Microfluidic-Based Capture and Release of Cancer-Derived Exosomes via Peptide–Nanowire Hybrid Interface. Lab Chip 2021, 21, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Radnaa, E.; Richardson, L.S.; Sheller-Miller, S.; Baljinnyam, T.; De Castro Silva, M.; Kumar Kammala, A.; Urrabaz-Garza, R.; Kechichian, T.; Kim, S.; Han, A.; et al. Extracellular Vesicle Mediated Feto-Maternal HMGB1 Signaling Induces Preterm Birth. Lab Chip 2021, 21, 1956–1973. [Google Scholar] [CrossRef] [PubMed]
- Lo, T.W.; Figueroa-Romero, C.; Hur, J.; Pacut, C.; Stoll, E.; Spring, C.; Lewis, R.; Nair, A.; Goutman, S.A.; Sakowski, S.A.; et al. Extracellular Vesicles in Serum and Central Nervous System Tissues Contain MicroRNA Signatures in Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2021, 14, 246. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gui, Y.; Liu, W.; Li, C.; Yang, Y. Precise Molecular Profiling of Circulating Exosomes Using a Metal–Organic Framework-Based Sensing Interface and an Enzyme-Based Electrochemical Logic Platform. Anal. Chem. 2022, 94, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Paisrisarn, P.; Yasui, T.; Zhu, Z.; Klamchuen, A.; Kasamechonchung, P.; Wutikhun, T.; Yordsri, V.; Baba, Y. Tailoring ZnO Nanowire Crystallinity and Morphology for Label-Free Capturing of Extracellular Vesicles. Nanoscale 2022, 14, 4484–4494. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-Based Analysis of Exosomal MRNA Mediating Drug Resistance in Glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef]
- Zhao, Z.; McGill, J.; Gamero-Kubota, P.; He, M. Microfluidic On-Demand Engineering of Exosomes towards Cancer Immunotherapy. Lab Chip 2019, 19, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Chen, X.; Niu, X.; Cai, Y.; Zhang, Q.; Chen, T.; Yang, H. Integrated Immunomagnetic Bead-Based Microfluidic Chip for Exosomes Isolation. Micromachines 2020, 11, 503. [Google Scholar] [CrossRef]
- Song, Z.; Mao, J.; Barrero, R.A.; Wang, P.; Zhang, F.; Wang, T. Development of a CD63 Aptamer for Efficient Cancer Immunochemistry and Immunoaffinity-Based Exosome Isolation. Molecules 2020, 25, 5585. [Google Scholar] [CrossRef]
- Huang, C.C.; Kuo, Y.H.; Chen, Y.S.; Huang, P.C.; Lee, G. Bin A Miniaturized, DNA-FET Biosensor-Based Microfluidic System for Quantification of Two Breast Cancer Biomarkers. Microfluid. Nanofluidics 2021, 25, 1–12. [Google Scholar] [CrossRef]
- Zhao, Y.; Fang, X.; Bai, M.; Zhang, J.; Yu, H.; Chen, F.; Zhao, Y. A Microfluidic Surface-Enhanced Raman Scattering (SERS) Sensor for MicroRNA in Extracellular Vesicles with Nucleic Acid-Tyramine Cascade Amplification. Chin. Chem. Lett. 2022, 33, 2101–2104. [Google Scholar] [CrossRef]
- Choi, Y.; Park, U.; Koo, H.J.; Park, J.S.; Lee, D.H.; Kim, K.; Choi, J. Exosome-Mediated Diagnosis of Pancreatic Cancer Using Lectin-Conjugated Nanoparticles Bound to Selective Glycans. Biosens. Bioelectron. 2021, 177, 112980. [Google Scholar] [CrossRef]
- Piffoux, M.; Silva, A.K.A.; Lugagne, J.B.; Hersen, P.; Wilhelm, C.; Gazeau, F. Extracellular Vesicle Production Loaded with Nanoparticles and Drugs in a Trade-off between Loading, Yield and Purity: Towards a Personalized Drug Delivery System. Adv. Biosyst. 2017, 1, 1700044. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, D.; Serra, M.; Filippi, D.; Zago, L.; Guglielmin, E.; Pierno, M.; Descroix, S.; Viovy, J.L.; Mistura, G. Controlling the Distance of Highly Confined Droplets in a Capillary by Interfacial Tension for Merging On-Demand. Lab Chip 2019, 19, 136–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salva, M.L.; Rocca, M.; Niemeyer, C.M.; Delamarche, E. Methods for Immobilizing Receptors in Microfluidic Devices: A Review. Micro Nano Eng. 2021, 11, 100085. [Google Scholar] [CrossRef]
- Kang, Y.T.; Kim, Y.J.; Bu, J.; Cho, Y.H.; Han, S.W.; Moon, B.I. High-Purity Capture and Release of Circulating Exosomes Using an Exosome-Specific Dual-Patterned Immunofiltration (ExoDIF) Device. Nanoscale 2017, 9, 13495–13505. [Google Scholar] [CrossRef]
- Nielsen, J.B.; Hanson, R.L.; Almughamsi, H.M.; Pang, C.; Fish, T.R.; Woolley, A.T. Microfluidics: Innovations in Materials and Their Fabrication and Functionalization. Anal. Chem. 2020, 92, 150–168. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Sahloul, S.; Orozaliev, A.; Do, V.Q.; Pham, V.S.; Martins, D.; Wei, X.; Levicky, R.; Song, Y.A. Microfluidic Electrokinetic Preconcentration Chips: Enhancing the Detection of Nucleic Acids and Exosomes. IEEE Nanotechnol. Mag. 2020, 14, 18–34. [Google Scholar] [CrossRef]
- Shiri, F.; Feng, H.; Petersen, K.E.; Sant, H.; Bardi, G.T.; Schroeder, L.A.; Merchant, M.L.; Gale, B.K.; Hood, J.L. Separation of U87 Glioblastoma Cell-Derived Small and Medium Extracellular Vesicles Using Elasto-Inertial Flow Focusing (a Spiral Channel). Sci. Rep. 2022, 12, 6146. [Google Scholar] [CrossRef] [PubMed]
- Leong, S.Y.; Ong, H.B.; Tay, H.M.; Kong, F.; Upadya, M.; Gong, L.; Dao, M.; Dalan, R.; Hou, H.W. Microfluidic Size Exclusion Chromatography (ΜSEC) for Extracellular Vesicles and Plasma Protein Separation. Small 2022, 18, 2104470. [Google Scholar] [CrossRef] [PubMed]
- Raju, D.; Bathini, S.; Badilescu, S.; Ouellette, R.J.; Ghosh, A.; Packirisamy, M. LSPR Detection of Extracellular Vesicles Using a Silver-PDMS Nano-Composite Platform Suitable for Sensor Networks. Enterp. Inf. Syst. 2018, 14, 532–541. [Google Scholar] [CrossRef]
- Wu, T.; Yang, Y.; Cao, Y.; Huang, Y.; Xu, L.P.; Zhang, X.; Wang, S. Enhanced Lateral Flow Assay with Double Conjugates for the Detection of Exosomes. Sci. China Chem. 2018, 61, 1423–1429. [Google Scholar] [CrossRef]
- Yokota, S.; Kuramochi, H.; Okubo, K.; Iwaya, A.; Tsuchiya, S.; Ichiki, T. Extracellular Vesicles Nanoarray Technology: Immobilization of Individual Extracellular Vesicles on Nanopatterned Polyethylene Glycol-Lipid Conjugate Brushes. PLoS ONE 2019, 14, e0224091. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Crow, J.; Lella, D.; Zhou, X.; Samuel, G.; Godwin, A.K.; Zeng, Y. Ultrasensitive Quantification of Tumor MRNAs in Extracellular Vesicles with an Integrated Microfluidic Digital Analysis Chip. Lab Chip 2018, 18, 3790–3801. [Google Scholar] [CrossRef]
- Zhou, Q.; Rahimian, A.; Son, K.; Shin, D.S.; Patel, T.; Revzin, A. Development of an Aptasensor for Electrochemical Detection of Exosomes. Methods 2016, 97, 88–93. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Zhang, R.Y.; Sun, N.; Smalley, M.; Wu, Z.; Zhou, A.; Chou, S.J.; Jan, Y.J.; Yang, P.; Bao, L.; et al. Bio-Inspired NanoVilli Chips for Enhanced Capture of Tumor-Derived Extracellular Vesicles: Toward Non-Invasive Detection of Gene Alterations in Non-Small Cell Lung Cancer. ACS Appl. Mater. Interfaces 2019, 11, 13973–13983. [Google Scholar] [CrossRef]
- Chen, W.; Cao, R.; Su, W.; Zhang, X.; Xu, Y.; Wang, P.; Gan, Z.; Xie, Y.; Li, H.; Qin, J. Simple and Fast Isolation of Circulating Exosomes with a Chitosan Modified Shuttle Flow Microchip for Breast Cancer Diagnosis. Lab Chip 2021, 21, 1759–1770. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.T.; Hadlock, T.; Jolly, S.; Nagrath, S. Extracellular Vesicles on Demand (EVOD) Chip for Screening and Quantification of Cancer-Associated Extracellular Vesicles. Biosens. Bioelectron. 2020, 168, 112535. [Google Scholar] [CrossRef] [PubMed]
- Chutvirasakul, B.; Nuchtavorn, N.; Suntornsuk, L.; Zeng, Y. Exosome Aggregation Mediated Stop-Flow Paper-Based Portable Device for Rapid Exosome Quantification. Electrophoresis 2020, 41, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular Vesicle Sizing and Enumeration by Nanoparticle Tracking Analysis. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, V.; Lucchetti, D.; Gatto, I.; Maiorana, A.; Marcantoni, M.; Maulucci, G.; Papi, M.; Pola, R.; De Spirito, M.; Sgambato, A. Dynamic Light Scattering for the Characterization and Counting of Extracellular Vesicles: A Powerful Noninvasive Tool. J. Nanoparticle Res. 2014, 16. [Google Scholar] [CrossRef]
- Welsh, J.A.; Holloway, J.A.; Wilkinson, J.S.; Englyst, N.A. Extracellular Vesicle Flow Cytometry Analysis and Standardization. Front. Cell Dev. Biol. 2017, 5, 78. [Google Scholar] [CrossRef]
- Ortega, F.G.; Piguillem, S.V.; Messina, G.A.; Tortella, G.R.; Rubilar, O.; Jiménez Castillo, M.I.; Lorente, J.A.; Serrano, M.J.; Raba, J.; Fernández Baldo, M.A. EGFR Detection in Extracellular Vesicles of Breast Cancer Patients through Immunosensor Based on Silica-Chitosan Nanoplatform. Talanta 2019, 194, 243–252. [Google Scholar] [CrossRef]
- Cimorelli, M.; Nieuwland, R.; Varga, Z.; van der Pol, E. Standardized Procedure to Measure the Size Distribution of Extracellular Vesicles Together with Other Particles in Biofluids with Microfluidic Resistive Pulse Sensing. PLoS ONE 2021, 16, e0249603. [Google Scholar] [CrossRef]
- Wang, S.; Khan, A.; Huang, R.; Ye, S.; Di, K.; Xiong, T.; Li, Z. Recent Advances in Single Extracellular Vesicle Detection Methods. Biosens. Bioelectron. 2020, 154, 112056. [Google Scholar] [CrossRef]
- Bordanaba-Florit, G.; Royo, F.; Kruglik, S.G.; Falcón-Pérez, J.M. Using Single-Vesicle Technologies to Unravel the Heterogeneity of Extracellular Vesicles. Nat. Protoc. 2021, 16, 3163–3185. [Google Scholar] [CrossRef]

| Working Principle | Retention Time | Output Sample (Quality and Quantity) | Simplicity | |
|---|---|---|---|---|
| Differential ultracentrifugation | particle size | - | - | ++ |
| Density gradient ultracentrifugation | particle density | -- | -- | ++ |
| Ultrafiltration | particle size | -- | - | + |
| Size-exclusion chromatography | particle size | -- | - | + |
| Field-flow fractionation | particle size | - | + | + |
| Precipitation-based | particle–polymer interaction | - | + | + |
| Immunoaffinity-based | antigen–antibody binding | - | + | + |
| Methods | Working Principle | |
|---|---|---|
| Physical: Passive | Filtration [38,39,40,41,42,43,44,45,150,151] | Micro-/nano- filtration process by porous membranes inside chip |
| Deterministic lateral displacement [46,60,61,62] | Particle distribution in size by lateral forces conveyed by ordered array of posts | |
| Inertial force [50,51,52,53,54,55,77] Viscoelastic force [67,68,69,70,71,72] | Imbalance of inertial forces or of shear forces in non-Newtonian viscoelastic fluid | |
| Physical: Active | Acoustofluidics [86,87,88,89,90,91,92,93,94,95,96,107] | Acoustic trapping by ultrasound waves |
| Electrokinetic force [98,99,100,101,102,103,104,105,106,107,152,153,154] | Charge separation by electric fields | |
| Chemical: Fixed support | Functionalized fixed support [108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,130,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180] | EV capture by specific antibodies on fixed substrate |
| Chemical: Floating Beads | Magnetic beads [125,126,127,128,129,130,131,132,133,134,135,181,182,183,184,185,186] | EV capture by specific antibodies on beads for magnetic manipulation |
| Polystyrene beads [80,99,124,137,139,140,141,187] | EV capture by specific antibodies on non-magnetic beads | |
| Magnetic nanoparticles [93,134,138,142,143,144,145,146,147,149,188] | EV capture or handling by specific antibodies on magnetic nanoparticles |
| Throughput | Output Sample (Quality and Quantity) | Possible Automation | Micro- Fabrication Simplicity | |
|---|---|---|---|---|
| Physical: Passive | ++ | - | + | - |
| Physical: Active | - | + | + | - |
| Chemical: Fixed support | + | ++ | ++ | + |
| Chemical: Floating Beads | + | ++ | -- | + |
| Proof of Concept | Clinical Sample | |||
|---|---|---|---|---|
| Starting Sample | % | Starting Sample | % | |
| Physical: Passive | Plasma [52,62] Urine [44,60,95] Cell culture [41,42,43,50,51,52,54,55,61,68,71,72,77,88,150,151,193,194] | 12 | Plasma [39,195] Serum [43,46,71] Blood [45,53,67,69] Urine [38,39,44,46] | 8 |
| Physical: Active | Plasma [98,102,103,106,152] Serum [103] Saliva [88] Cell culture [86,89,91,98,99,100,104,105,107,153,154] | 12 | Plasma [87,91,94] Blood [90,101] Urine [91,92,95] | 5 |
| Chemical: Fixed support | Plasma [87,173] Serum [122,168] Cell culture [109,113,122,155,156,157,158,160,161,162,164,165,166,167,172,174,175,176,177,179,180,196,197,198,199,200] | 24 | Plasma [108,114,119,120,123,155,163,171,173,201] Serum [110,117,118,123,130,178,202,203] Blood [170] Urine [115,159,169] | 14 |
| Chemical: Floating beads | Plasma [130] Serum [130,183] Cell culture [99,137,139,140,141,143,147,182,186,188,193,204] | 11 | Plasma [80,124,125,126,127,129,132,135,138,144,185] Serum [131,134,145,147,148,181] Blood [133,142,146] Urine [128] | 14 |
| TOTAL | 59 | 41 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meggiolaro, A.; Moccia, V.; Brun, P.; Pierno, M.; Mistura, G.; Zappulli, V.; Ferraro, D. Microfluidic Strategies for Extracellular Vesicle Isolation: Towards Clinical Applications. Biosensors 2023, 13, 50. https://doi.org/10.3390/bios13010050
Meggiolaro A, Moccia V, Brun P, Pierno M, Mistura G, Zappulli V, Ferraro D. Microfluidic Strategies for Extracellular Vesicle Isolation: Towards Clinical Applications. Biosensors. 2023; 13(1):50. https://doi.org/10.3390/bios13010050
Chicago/Turabian StyleMeggiolaro, Alessio, Valentina Moccia, Paola Brun, Matteo Pierno, Giampaolo Mistura, Valentina Zappulli, and Davide Ferraro. 2023. "Microfluidic Strategies for Extracellular Vesicle Isolation: Towards Clinical Applications" Biosensors 13, no. 1: 50. https://doi.org/10.3390/bios13010050







