A Multi-Drug Concentration Gradient Mixing Chip: A Novel Platform for High-Throughput Drug Combination Screening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fabrication of Microfluidic Chips
2.2. Numerical Simulation
2.3. Chip Operation
2.4. Cell Culture and Loading
2.5. Image Acquisition and Data Analysis
3. Results
3.1. Design and Characterization of Microfluidic Chips
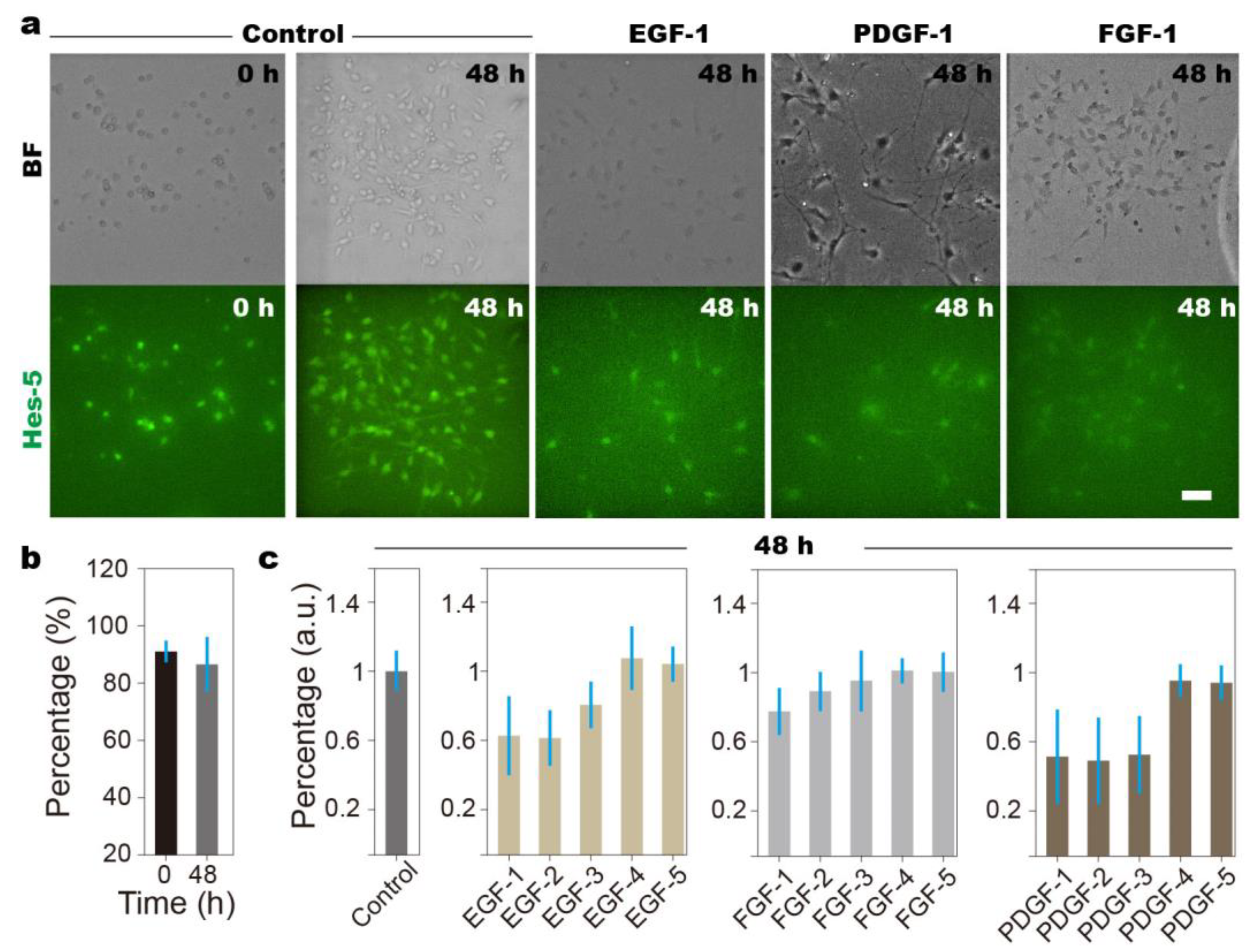
3.2. Overdose of Growth Factors Leads to NSCs’ Diminished Stemness
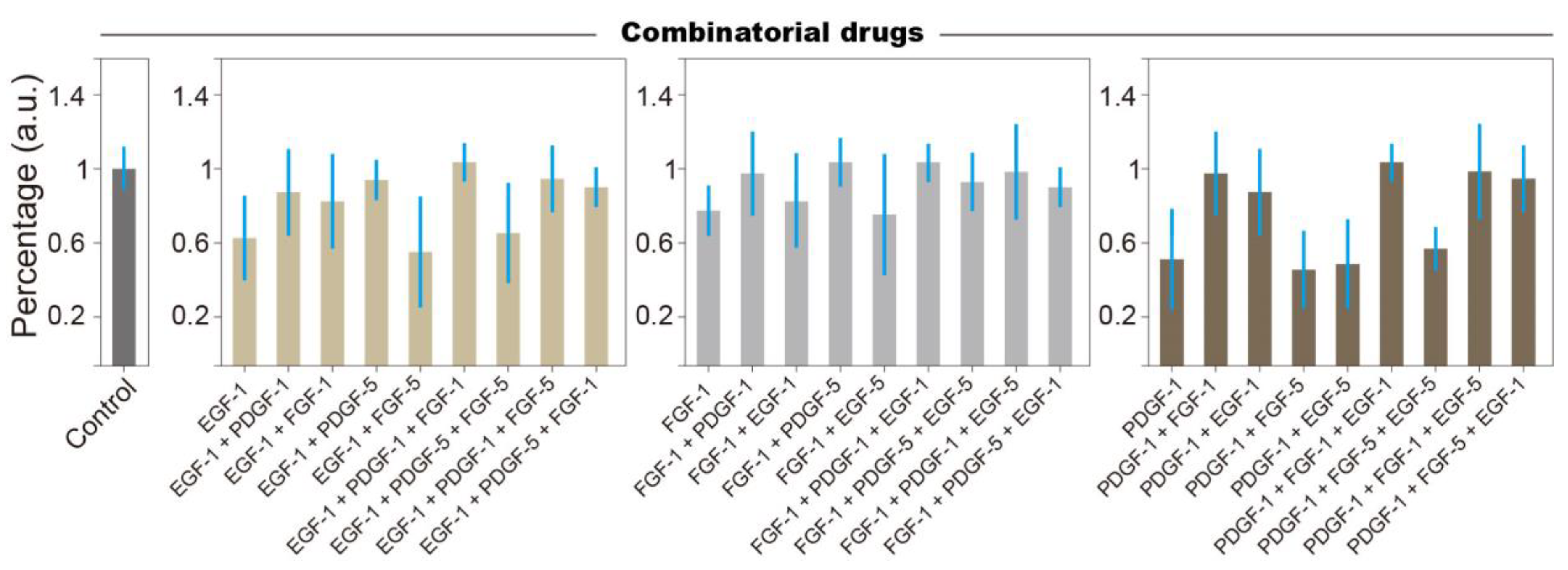
3.3. Combinatorial Treatment Reveals Logic Rules of Growth Factor Affecting NSCs Stemness
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lawler, S.E.; Chiocca, E.A. Oncolytic Virus-Mediated Immunotherapy: A Combinatorial Approach for Cancer Treatment. J. Clin. Oncol. 2015, 33, 2812–2814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, G.; Zhong, K.; Chen, Y.; Yang, N.; Lu, Q.; Yuan, B.; Wang, Z.; Li, H.; Guo, L.; et al. A drug screening to identify novel combinatorial strategies for boosting cancer immunotherapy efficacy. J. Transl. Med. 2023, 21, 23. [Google Scholar] [CrossRef] [PubMed]
- Vilgelm, A.E.; Johnson, D.B.; Richmond, A. Combinatorial approach to cancer immunotherapy: Strength in numbers. J. Leukoc. Biol. 2016, 100, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Normann, L.S.; Haugen, M.H.; Hongisto, V.; Aure, M.R.; Leivonen, S.K.; Kristensen, V.N.; Tahiri, A.; Engebraaten, O.; Sahlberg, K.K.; Mælandsmo, G.M. High-throughput screen in vitro identifies dasatinib as a candidate for combinatorial treatment with HER2-targeting drugs in breast cancer. PLoS ONE 2023, 18, e0280507. [Google Scholar] [CrossRef] [PubMed]
- Flobak, Å.; Niederdorfer, B.; Nakstad, V.T.; Thommesen, L.; Klinkenberg, G.; Lægreid, A. A high-throughput drug combination screen of targeted small molecule inhibitors in cancer cell lines. Sci. Data 2019, 6, 237. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Vilar, S.; Tatonetti, N.P. High-throughput methods for combinatorial drug discovery. Sci. Transl. Med. 2013, 5, 205rv1. [Google Scholar] [CrossRef]
- Held, M.A.; Langdon, C.G.; Platt, J.T.; Graham-Steed, T.; Liu, Z.; Chakraborty, A.; Bacchiocchi, A.; Koo, A.; Haskins, J.W.; Bosenberg, M.W.; et al. Genotype-Selective Combination Therapies for Melanoma Identified by High-Throughput Drug Screening. Cancer Discov. 2013, 3, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Dang, D.; Wang, W.; Wang, Y.; Liu, L. Concentration optimization of combinatorial drugs using Markov chain-based models. BMC Bioinform. 2021, 22, 451. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Wang, L.; Xu, H. Application of kriging models for a drug combination experiment on lung cancer. Stat. Med. 2019, 38, 236–246. [Google Scholar] [CrossRef]
- Lin, X.; Li, X.; Lin, X. A Review on Applications of Computational Methods in Drug Screening and Design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef]
- Schuster, B.; Junkin, M.; Kashaf, S.S.; Romero-Calvo, I.; Kirby, K.; Matthews, J.; Weber, C.R.; Rzhetsky, A.; White, K.P.; Tay, S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 2020, 11, 5271. [Google Scholar] [CrossRef]
- Wan, L.; Yin, J.; Skoko, J.; Schwartz, R.; Zhang, M.; LeDuc, P.R.; Neumann, C.A. 3D Collagen Vascular Tumor-on-a-Chip Mimetics for Dynamic Combinatorial Drug Screening. Mol. Cancer Ther. 2021, 20, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Petreus, T.; Cadogan, E.; Hughes, G.; Smith, A.; Pilla Reddy, V.; Lau, A.; O’Connor, M.J.; Critchlow, S.; Ashford, M.; Oplustil O’Connor, L. Tumour-on-chip microfluidic platform for assessment of drug pharmacokinetics and treatment response. Commun. Biol. 2021, 4, 1001. [Google Scholar] [CrossRef]
- Chen, L.; Ji, Y.; Li, A.; Liu, B.; Shen, K.; Su, R.; Ma, Z.; Zhang, W.; Wang, Q.; Zhu, Y.; et al. High-throughput drug screening identifies fluoxetine as a potential therapeutic agent for neuroendocrine prostate cancer. Front Oncol. 2023, 13, 1085569. [Google Scholar] [CrossRef]
- Oudebrouckx, G.; Goossens, J.; Bormans, S.; Vandenryt, T.; Wagner, P.; Thoelen, R. Integrating Thermal Sensors in a Microplate Format: Simultaneous Real-Time Quantification of Cell Number and Metabolic Activity. ACS Appl Mater Interfaces 2022, 14, 2440–2451. [Google Scholar] [CrossRef] [PubMed]
- Dasovich, M.; Zhuo, J.; Goodman, J.A.; Thomas, A.; McPherson, R.L.; Jayabalan, A.K.; Busa, V.F.; Cheng, S.J.; Murphy, B.A.; Redinger, K.R.; et al. High-Throughput Activity Assay for Screening Inhibitors of the SARS-CoV-2 Mac1 Macrodomain. ACS Chem Biol. 2022, 17, 17–23. [Google Scholar] [CrossRef]
- Pham, N.; Radajewski, D.; Round, A.; Brennich, M.; Pernot, P.; Biscans, B.; Bonneté, F.; Teychené, S. Coupling High Throughput Microfluidics and Small-Angle X-ray Scattering to Study Protein Crystallization from Solution. Anal. Chem. 2017, 89, 2282–2287. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, I.; Radajewski, D.; Charton, S.; Phamvan, N.; Brennich, M.; Pernot, P.; Bonneté, F.; Teychené, S. Innovative High-Throughput SAXS Methodologies Based on Photonic Lab-on-a-Chip Sensors: Application to Macromolecular Studies. Sensors 2017, 17, 1266. [Google Scholar] [CrossRef]
- Ansari, M.I.H.; Hassan, S.; Qurashi, A.; Khanday, F.A. Microfluidic-integrated DNA nanobiosensors. Biosens. Bioelectron. 2016, 85, 247–260. [Google Scholar] [CrossRef]
- Lou, C.; Yang, H.; Hou, Y.; Huang, H.; Qiu, J.; Wang, C.; Sang, Y.; Liu, H.; Han, L. Microfluidic Platforms for Real-Time in Situ Monitoring of Biomarkers for Cellular Processes. Adv. Mater. 2023, 36, e2307051. [Google Scholar] [CrossRef]
- Carr, J.A.; Parashar, A.; Gibson, R.; Robertson, A.P.; Martin, R.J.; Pandey, S. A microfluidic platform for high-sensitivity, real-time drug screening on C. elegans and parasitic nematodes. Lab Chip 2011, 11, 2385–2396. [Google Scholar] [CrossRef]
- Zuieva, A.; Can, S.; Boelke, F.; Reuter, S.; Schattscheider, S.; Töpfer, E.; Westphal, A.; Mrowka, R.; Wölfl, S. Real-time monitoring of immediate drug response and adaptation upon repeated treatment in a microfluidic chip system. Arch. Toxicol. 2022, 96, 1483–1487. [Google Scholar] [CrossRef]
- Tay, S.; Hughey, J.J.; Lee, T.K.; Lipniacki, T.; Quake, S.R.; Covert, M.W. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 2010, 466, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tu, H.L.; Jia, G.; Mukhtar, T.; Taylor, V.; Rzhetsky, A.; Tay, S. Ultra-multiplexed analysis of single-cell dynamics reveals logic rules in differentiation. Sci. Adv. 2019, 5, eaav7959. [Google Scholar] [CrossRef]
- Han, S.-I.; Han, K.-H.; Frazier, A.B.; Ferrance, J.P.; Landers, J.P. An automated micro-solid phase extraction device involving integrated \high-pressure microvalves for genetic sample preparation. Biomed. Microdevices 2009, 11, 935–942. [Google Scholar] [CrossRef]
- Ansede, J.H.; Thakker, D.R. High-throughput screening for stability and inhibitory activity of compounds toward cytochrome P450-mediated metabolism. J. Pharm. Sci. 2004, 93, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Konkankit, C.C.; Vaughn, B.A.; MacMillan, S.N.; Boros, E.; Wilson, J.J. Combinatorial Synthesis to Identify a Potent, Necrosis-Inducing Rhenium Anticancer Agent. Inorg. Chem. 2019, 58, 3895–3909. [Google Scholar] [CrossRef]
- Zhang, Y.; Yazid, N.B.M.; Ho, P.Y.; Hu, X.; Chen, S.; Vasoo, S.; Kanitthamniyom, P. DropCarba—An automated magnetic digital microfluidic platform for rapid phenotypic testing of carbapenemase-producing Gram-negative bacilli. Biosens. Bioelectron. 2023, 225, 115099. [Google Scholar] [CrossRef]
- Goto, M.; Hojo, M.; Ando, M.; Kita, A.; Kitagawa, M.; Ohtsuka, T.; Kageyama, R.; Miyamoto, S. Hes1 and Hes5 are required for differentiation of pituicytes and formation of the neurohypophysis in pituitary development. Brain Res. 2015, 1625, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Manning, C.S.; Biga, V.; Boyd, J.; Kursawe, J.; Ymisson, B.; Spiller, D.G.; Sanderson, C.M.; Galla, T.; Rattray, M.; Papalopulu, N. Quantitative single-cell live imaging links HES5 dynamics with cell-state and fate in murine neurogenesis. Nat. Commun. 2019, 10, 2835. [Google Scholar] [CrossRef]
- Unger, M.A.; Chou, H.P.; Thorsen, T.; Scherer, A.; Quake, S.R. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science 2000, 288, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Giachino, C.; Basak, O.; Taylor, V. Isolation and manipulation of mammalian neural stem cells in vitro. Methods Mol. Biol. 2009, 482, 143–158. [Google Scholar] [PubMed]
- Sykova, E.; Forostyak, S. Stem cells in regenerative medicine. Laser Ther. 2013, 22, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Behnan, J.; Grieg, Z.; Joel, M.; Ramsness, I.; Stangeland, B. Neuroepigenetics Gene knockdown of CENPA reduces sphere forming ability and stemness of glioblastoma initiating cells. Neuroepigenetics 2016, 7, 6–18. [Google Scholar] [CrossRef]
- Shao, J.; Wu, L.; Wu, J.; Zheng, Y.; Zhao, H.; Jin, Q.; Zhao, J. Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress. Lab Chip 2009, 9, 3118–3125. [Google Scholar] [CrossRef] [PubMed]
- Mehling, M.; Tay, S. Microfluidic cell culture. Curr. Opin. Biotechnol. 2014, 25, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Nakamura, H.; Goto, T.; Uchida, W.; Uozumi, T.; Nishizawa, D.; Shinha, K.; Sakagami, J.; Doi, K. Standalone cell culture microfluidic device-based microphysiological system for automated cell observation and application in nephrotoxicity tests. Lab Chip 2024, 24, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Prentice, H.; Oliveira, J.; Madziva, N.; Warkiani, M.E.; Hamel, J.F.P.; Han, J. Microfluidic Cell Retention Device for Perfusion of Mammalian Suspension Culture. Sci. Rep. 2017, 7, 6703. [Google Scholar] [CrossRef] [PubMed]
- Van Midwoud, P.M.; Merema, M.T.; Verpoorte, E.; Groothuis, G.M. Microfluidics enables small-scale tissue-based drug metabolism studies with scarce human tissue. J. Lab. Autom. 2011, 16, 468–476. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, W.; Jiang, X. Cell-Based Assays on Microfluidics for Drug Screening. ACS Sens. 2019, 4, 1465–1475. [Google Scholar] [CrossRef]
- Kohl, Y.; Biehl, M.; Spring, S.; Hesler, M.; Ogourtsov, V.; Todorovic, M.; Owen, J.; Elje, E.; Kopecka, K.; Moriones, O.H.; et al. Microfluidic In Vitro Platform for (Nano)Safety and (Nano)Drug Efficiency Screening. Small 2021, 17, e2006012. [Google Scholar] [CrossRef] [PubMed]
- Yeo, L.Y.; Chang, H.C.; Chan, P.P.; Friend, J.R. Microfluidic devices for bioapplications. Small 2011, 7, 12–48. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Cao, R.; Che, B.; Sun, D.; Tang, Y.; Jiang, L.; Bai, Q.; Liu, Y.; Morozova-Roche, L.A.; Zhang, C. Proinflammatory S100A9 Regulates Differentiation and Aggregation of Neural Stem Cells. ACS Chem. Neurosci. 2020, 11, 3549–3556. [Google Scholar] [CrossRef] [PubMed]
- Gan, D.; Cheng, W.; Ke, L.; Sun, A.R.; Jia, Q.; Chen, J.; Xu, Z.; Xu, J.; Zhang, P. Biphasic Effect of Pirfenidone on Angiogenesis. Front. Pharmacol. 2022, 12, 804327. [Google Scholar] [CrossRef] [PubMed]
- Al-Humadi, N.H.; Ma, J.K.; Lewis, D.M.; Ma, J.Y.; Barger, M.W.; Siegel, P.D. Dose-dependent thiol and immune responses to ovalbumin challenge in Brown Norway rats. Toxicol. Ind. Health 2002, 18, 343–352. [Google Scholar] [CrossRef]
- Jackson, E.L.; Garcia-Verdugo, J.M.; Gil-Perotin, S.; Roy, M.; Quinones-Hinojosa, A.; VandenBerg, S.; Alvarez-Buylla, A. PDGFRα-Positive B Cells Are Neural Stem Cells in the Adult SVZ that Form Glioma-like Growths in Response to Increased PDGF Signaling. Neuron 2006, 51, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.G.; Wang, Y.X.; Wang, H.J.; Bao, M.S.; Wang, Z.H.; Ge, X.; Wang, F.C.; Zhou, J.S.; Lü, H.Z. PDGF-AA Mediates B104CM-Induced Oligodendrocyte Precursor Cell Differentiation of Embryonic Neural Stem Cells Through Erk, PI3K, and p38 Signaling. J. Mol. Neurosci. 2011, 46, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, A.; Klinge, A.; Nedergaard, J.; Siemen, D. Regulation of the Activity of 27 pS Nonselective Cation Channels in Excised Membrane Patches from Rat Brown-Fat Cells. Cell. Physiol. Biochem. 1998, 8, 231–245. [Google Scholar] [CrossRef]
- Mondal, D.; Pradhan, L.; Larussa, V. Signal Transduction Pathways Involved in the Lineage-Specific Differentiation of NSCs: Can the Knowledge Gained from Blood be Used in the Brain? Cancer Investig. 2004, 22, 925–943. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, J.; Feng, Y.; Sun, Y.; Yi, R.; Tian, J.; Zhao, W.; Sun, D.; Zhang, C. A Multi-Drug Concentration Gradient Mixing Chip: A Novel Platform for High-Throughput Drug Combination Screening. Biosensors 2024, 14, 212. https://doi.org/10.3390/bios14050212
Fu J, Feng Y, Sun Y, Yi R, Tian J, Zhao W, Sun D, Zhang C. A Multi-Drug Concentration Gradient Mixing Chip: A Novel Platform for High-Throughput Drug Combination Screening. Biosensors. 2024; 14(5):212. https://doi.org/10.3390/bios14050212
Chicago/Turabian StyleFu, Jiahao, Yibo Feng, Yu Sun, Ruiya Yi, Jing Tian, Wei Zhao, Dan Sun, and Ce Zhang. 2024. "A Multi-Drug Concentration Gradient Mixing Chip: A Novel Platform for High-Throughput Drug Combination Screening" Biosensors 14, no. 5: 212. https://doi.org/10.3390/bios14050212




