The Heme-Based Oxygen-Sensor Phosphodiesterase Ec DOS (DosP): Structure-Function Relationships
Abstract
:1. Introduction

2. Catalytic Activities



2.1. Catalytic Activity toward c-AMP
2.1.1. Heme Redox-Dependent PDE Activity toward c-di-AMP
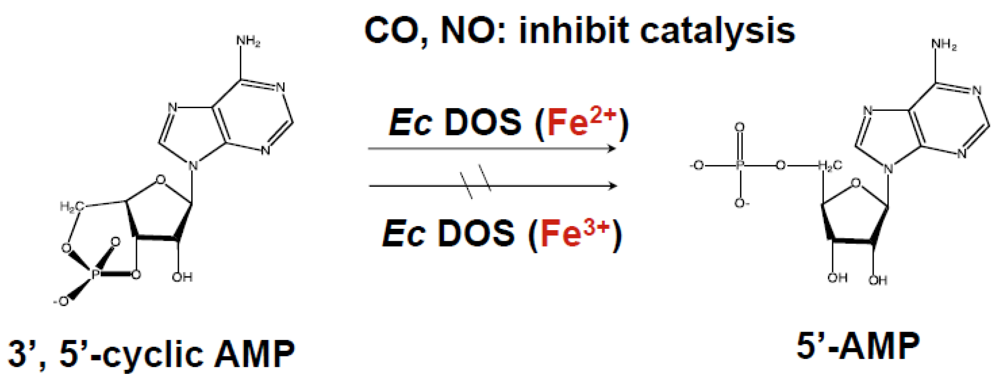

2.1.2. Removal of Heme or Truncation of the Heme-Bound PAS Domain from Ec DOS Does Not Influence the Catalytic Activity toward c-AMP
2.1.3. Addition of Ec DOS-PAS-A to the Full-Length Ec DOS Enzyme Regulates Catalysis

2.1.4. Issues of Catalysis toward c-AMP and Redox Sensor
2.2. Catalysis of c-di-GMP
2.2.1. O2 (NO/CO) Binding to the Heme Fe(II) Complex Enhances Catalysis via Dissociation of the Axial Ligand, M95

2.2.2. Addition of the Exogenous Ligands, Cyanide and Imidazole, to Ec DOS-Heme Fe(III) Stimulates Catalysis
2.2.3. The Heme Iron Complex Is Not Essential for Intrinsic Catalytic Activity
2.2.4. Catalysis with Mn2+ Proceeds without Gas Molecules
2.2.5. Interactions between Hydrogen Sulfide and the Wild Type and R97 Mutant Proteins

3. Protein Structures
3.1. X-ray Crystal Structures: Ligand- and Redox-Dependent Conformational Changes
3.2. Domain Structures
4. Physicochemical Characterizations
4.1. The Kd Values for O2 and CO Binding to Ec DOS are Very High
| Reduction potential | kon | Kd | kox | Ref. | |
|---|---|---|---|---|---|
| (mV vs. SHE) | (×10−3 μM−1s−1) | (μM) | (min−1) | ||
| WT | 45–70 | 37–81 | 20–21 | 0.0053 | [22,33,46,49] |
| D40A | 95 | 0.051 | [41] | ||
| D40N | 114 | 0.033 | |||
| H83A | 0.01 | [42] | |||
| N84A | 0.0015 | ||||
| R85A | 0.0026 | ||||
| E86A | 0.0054 | ||||
| K89A | 0.0043 | ||||
| R91A | 0.54 | ||||
| E93A | 0.0057 | ||||
| S96A | 0.0063 | ||||
| M95A | −26 | >1,000 | <0.73 | 0.0013 | [46,50] |
| M95H | −122 | >1,000 | <0.79 | 0.016 | |
| M95I | 160 | 1.4 | [51] | ||
| M95L | −1 | >1,000 | <0.45 | 0.0017 | [46,50] |
| R97A | 43 | 76 | >9.5 | [33] | |
| R97E | 40 | >45 | |||
| R97I | 49 | 155 | 500 | 0.16 | |
| L99T | 23 | 49 | 0.049 | [49] | |
| L99F | 24 | 75 | 0.37 | ||
| F113L | 29 | >200 | 0.00068 | [42,52] | |
| F113T | 43 | 78 | 0.018 | ||
| F113Y | −27 | 26 | 0.039 | ||
| L115T | 35 | 55 | 0.065 | [49] | |
| L115F | 0.33 |
4.2. O2 and CO Binding Kinetics and the Stability of the Heme Fe(II)-O2 Complex are Substantially Altered by Mutations at M95 and R97
4.3. Cyanide and Imidazole Binding is Influenced by Mutations at M95 and R97
4.4. Reduction Potential Values are Important for O2-Regulated Catalysis
4.5. Infrared Spectra are Not Changed by Mutations at M95
4.6. Resonance Raman Spectra: Role of Hydrogen-Bonding Networks Involving M95, R97, W53, Y126, Heme Propionate, and Heme Vinyl in Signal Transduction
4.7. Pulse Radiolysis: Allosterically Regulated O2 Binding
4.8. Ultrafast Ligand Rebinding: Allosterically Regulated Catalysis and Involvement of Met95 in Signal Transduction
5. Site-Directed Mutations at Sites other than M95 and R97
5.1. Mutations at L99, F113, and L115 on the Heme Distal Side Change Autoxidation Rate Constants and Reduction Potentials
5.2. Mutations at D40 at the Heme Proximal Side Abolish Catalytic Activity toward c-AMP and Change Autoxidation Rate Constants and Reduction Potential Values
5.3. Amino Acids in the F-G Loop are Crucial for Heme Affinity and Autoxidation Rate Constant (i.e., Stability of the Heme Fe(II)-O2 Complex)
5.4. Fluorescence Spectra Suggest that W53 and W110 are Located Near the Heme and on the Protein Surface, Respectively
6. Genetic Studies
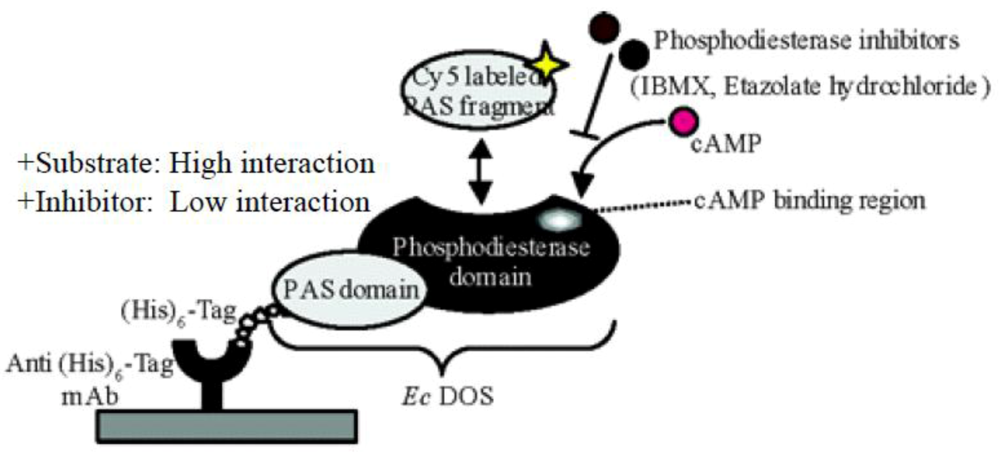
7. Application of a Protein Microarray to Study Ec DOS Interactions
8. Conclusions
Acknowledgements
| Abbreviations | |
|---|---|
| c-di-GMP | cyclic diguanylate monophosphate, bis(3'-5')-cyclic dimeric guanosine monophosphate |
| CRP | c-AMP receptor protein; c-AMP-dependent transcriptional regulator |
| DGC | diguanylate cyclase; synthesis of c-di-GMP |
| Ec DOS | E. coli Direct Oxygen Sensor; heme-based oxygen-sensor phosphodiesterase from E. coli, also designated Ec DosP |
| Ec DOS-heme Fe(II) | full-length Ec DOS containing the heme Fe(II) complex |
| Ec DOS-heme Fe(III) | full-length Ec DOS containing the heme Fe(III) complex |
| Ec DOS-PAS-A | isolated heme-bound N-terminal domain containing a PAS structure |
| Ec DOS-PAS-A-heme Fe(II) | Ec DOS-PAS-A containing the heme Fe(II) complex |
| Ec DOS-PAS-A-heme Fe(III) | Ec DOS-PAS-A containing the heme Fe(III) complex |
| Ec DosC | heme-based oxygen sensor diguanylate cyclase from E. coli, also designated YddV |
| Ec DosP | heme-based oxygen-sensor phosphodiesterase from E. coli, also designated Ec DOS |
| Hb | Hemoglobin |
| heme Fe(II) | protoporphyrin IX-Fe(II) complex |
| heme Fe(III) | protoporhyrin IX-Fe(III) complex, or hemin |
| Kd | equilibrium dissociation constant |
| koff | dissociation rate constant |
| kon | association rate constant |
| l-di-GMP | linear diguanylate monophosphate, pGpG |
| Mb | Myoglobin |
| PAS | an acronym formed from Per (Drosophila period clock protein)- Arnt (vertebrate aryl hydrocarbon receptor nuclear translocator)- Sim (Drosophila single-minded protein) |
| PDE | phosphodiesterase; linearization of c-di-GMP |
| RR spectroscopy | resonance Raman spectroscopy |
| WT | wild type |
| YddV | heme-based oxygen sensor diguanylate cyclase from E. coli, also designated Ec DosC |
Conflict of Interest
References
- Igarashi, J.; Kitanishi, K.; Shimizu, T. Emerging role of heme as a signal and the gas-sensing site: Heme-sensing and gas-sensing proteins. In Handbook of Porphyrin Science; Kadish, K.M., Smith, K.M., Guilard, R., Eds.; World Scientific: Hackensack, NJ, USA, 2011; Volume 15, pp. 399–460. [Google Scholar]
- Uchida, T.; Kitagawa, T. Mechanism for transduction of the ligand-binding signal in heme-based gas sensory proteins revealed by resonance Raman spectroscopy. Acc. Chem. Res. 2005, 38, 662–670. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Gonzalez, G. Heme-based sensors: Defining characteristics, recent developments, and regulatory hypothesis. J. Inorg. Biochem. 2005, 99, 1–22. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Gonzalez, G. Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 2004, 96, 774–783. [Google Scholar] [CrossRef]
- Sasakura, Y.; Yoshimura-Suzuki, T.; Kurokawa, H.; Shimizu, T. Structure-function relationships of Ec DOS, a heme-regulated phosphodiesterase from Escherichia coli. Acc. Chem. Res. 2006, 39, 37–43. [Google Scholar] [CrossRef]
- Aono, S. Novel bacterial gas sensor proteins with transition metal-containing prothethic groups as active sites. Antioxid. Redox Sign. 2012, 16, 678–686. [Google Scholar] [CrossRef]
- Igarashi, J.; Kitanishi, K.; Martinkova, M.; Murase, M.; Iizuka, A.; Shimizu, T. The roles of thiolate-heme proteins, other than the P450 cytochromes, in the regulation of heme-sensor proteins. Acta Chim. Slov. 2008, 55, 67–74. [Google Scholar]
- Taylor, B.L.; Zhulin, I.B. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999, 63, 479–506. [Google Scholar]
- McIntosh, B.E.; Hogenesch, J.B.; Bradfield, C.A. Mammalian Per-Arnt-Sim proteins in environmental adaption. Ann. Rev. Physiol. 2010, 72, 625–645. [Google Scholar]
- Henry, J.T.; Crosson, S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Ann. Rev. Microbiol. 2011, 65, 261–286. [Google Scholar] [CrossRef]
- Tuckerman, J.R.; Gonzalez, G.; Sousa, E.H.S.; Wan, X.; Saito, J.A.; Alam, M.; Gilles-Gonzalez, M.A. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 2009, 48, 9764–9774. [Google Scholar]
- Tuckerman, J.R.; Gozalez, G.; Gilles-Gonzalez, M.A. Cyclic-di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J. Mol. Biol. 2011, 407, 633–639. [Google Scholar] [CrossRef]
- Park, H.; Suquet, C.; Savenkova, M.I.; Satterlee, J.D.; Kang, C. Cloning, purification, crystallizationand preliminary X-ray analysis of DOS heme domain, a new heme oxygen sensor in Escherichia coli. Acta Cryst. 2002, D58, 1504–1506. [Google Scholar]
- Suquet, C.; Savankova, M.; Satterlee, J.D. Recombinant PAS-heme domains of oxygen sensing proteins: High level production and physical characterization. Protein Express. Purif. 2005, 42, 182–193. [Google Scholar] [CrossRef]
- Green, J.; Crack, J.C.; Thomson, A.J.; LeBrun, N.E. Bacterial sensors of oxygen. Curr. Opion Microbiol. 2009, 12, 145–151. [Google Scholar]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef]
- Schirmer, T.; Jenal, U. Structure and mechanistic determinants of c-di-GMP signaling. Nat. Rev. Microbiol. 2009, 7, 724–735. [Google Scholar] [CrossRef]
- Römling, U.; Gomelsky, M.; Galperin, M.Y. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 2005, 57, 629–639. [Google Scholar] [CrossRef]
- Kitanishi, K.; Kobayashi, K.; Kawamura, Y.; Ishigami, I.; Ogura, T.; Nakajima, K.; Igarashi, J.; Tanaka, A.; Shimizu, T. Important roles of Tyr43 at the putative heme distal side in the oxygen recognition and stability of the Fe(II)-O2 complex of YddV, a globin-coupled heme-based oxygen sensor diguanylate cyclase. Biochemistry 2010, 49, 10381–10393. [Google Scholar] [CrossRef]
- Delgado-Nixon, V.M.; Gonzalez, G.; Gilles-Gonzalez, M.A. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 2000, 39, 2685–2691. [Google Scholar] [CrossRef]
- Yoshimura, T.; Sagami, I.; Sasakura, Y.; Shimizu, T. Relationships between heme incorporation, tetramer formation and catalysis of a heme-regulated phosphodiesterase from Escherichia coli: A study of deletion and site-directed mutants. J. Biol. Chem. 2003, 278, 53105–53111. [Google Scholar]
- Sasakura, Y.; Hirata, S.; Sugiyama, S.; Suzuki, S.; Taguchi, S.; Watanabe, M.; Matsui, T.; Sagami, I.; Shimizu, T. Characterization of a direct oxygen sensor heme protein from E. coli: Effects of the heme redox states and mutations at the heme binding site on catalysis and structure. J. Biol. Chem. 2002, 277, 23821–23827. [Google Scholar] [CrossRef]
- Chang, A.L.; Tuckerman, J.R.; Gonzalez, G.; Mayer, R.; Weinhouse, H.; Volman, G.; Amikam, D.; Benziman, M.; Gilles-Gonzalez, M.A. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 2001, 40, 3420–3426. [Google Scholar] [CrossRef]
- Takahashi, H.; Shimizu, T. Phosphodiesterase activity of Ec DOS, a heme-regulated enzyme from Escherichia coli, toward 3', 5'-cyclic diguanylic acid is obviously enhanced by O2 and CO binding. Chem. Lett. 2006, 35, 970–971. [Google Scholar] [CrossRef]
- Tanaka, A.; Takahashi, H.; Shimizu, T. Critical role of the heme axial ligand, Met95, in locking catalysis of the phosphodiesterase from Escherichia coli (Ec DOS) toward cyclic diGMP. J. Biol. Chem. 2007, 282, 21301–21307. [Google Scholar] [CrossRef]
- Tanaka, A.; Shimizu, T. Ligand binding to the Fe(III)-protoporphyrin IX complex of phosphodiesterase from Escherichia coli (Ec DOS) markedly enhances catalysis of cyclic di-GMP: Roles of Met95, Arg97, and Phe113 of the putative heme distal side in catalytic regulation and ligand binding. Biochemistry 2008, 47, 13438–13446. [Google Scholar] [CrossRef]
- Kurokawa, H.; Lee, D.S.; Watanabe, M.; Sagami, I.; Mikami, B.; Raman, C.S.; Shimizu, T. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J. Biol. Chem. 2004, 279, 20186–20193. [Google Scholar]
- Park, H.; Suquet, C.; Satterlee, J.D.; Kang, C. Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli DOS heme domain (Ec DOSH). Biochemistry 2004, 43, 2738–2746. [Google Scholar] [CrossRef]
- Green, J.; Paget, M.S. Bacterial redox sensors. Nat. Rev. Microbiol. 2004, 2, 954–966. [Google Scholar] [CrossRef]
- Sasakura, Y.; Kanda, K.; Yoshimura-Suzuki, Y.; Matsui, T.; Fukuzono, S.; Han, M.H.; Shimizu, T. Protein microarray system for detecting protein–protein interactions using an anti-His-tag antibody and fluorescence scanning: Effects of the heme redox state on protein–protein interactions of heme-regulated phosphodiesterase from Escherichia coli. Anal. Chem. 2004, 76, 6521–6527. [Google Scholar] [CrossRef]
- Sasakura, Y.; Kanda, K.; Yoshimura-Suzuki, T.; Matsui, T.; Fukuzono, S.; Shimizu, T. Investigation of the relationship between protein–protein interaction and catalytic activity of a heme-regulated phosphodiesterase from Escherichia coli (Ec DOS) by protein microarray. Biochemistry 2005, 44, 9598–9605. [Google Scholar]
- Imamura, R.; Yamanaka, K.; Ogura, T.; Hiraga, S.; Fujita, N.; Ishihama, A.; Niki, H. Identification of the cpdA gene encoding cyclic 3',5'-adenosine monophosphate phosphodiesterase in Escherichia coli. J. Biol. Chem. 1996, 271, 25423–25429. [Google Scholar]
- Ishitsuka, Y.; Araki, Y.; Tanaka, A.; Igarashi, J.; Ito, O.; Shimizu, T. Arg97 at the heme-distal side of the isolated heme-bound PAS domain of a heme-based oxygen sensor from Escherichia coli (Ec DOS) plays critical roles in autooxidation and binding of gases, particularly O2. Biochemistry 2008, 47, 8874–8884. [Google Scholar]
- Voet, D.; Voet, J.G. Biochemistry, 3rd ed.; John Wiley & Sons: New York, NY, USA, 2004. [Google Scholar]
- Honjo, A.; Takahashi, H.; Sekimoto, M.; Igarashi, J.; Shimizu, T. Novel effects of Mn2+ on the catalytic enhancement of the inactive form of the Fe(III) heme-bound Ec DOS, a heme-regulated phosphodiesterase from Escherichia coli. In Gas Sensors: Developments, Efficacy and Safety; Qiu, X., Ed.; Nova Science: Hauppauge, NY, USA, 2012; pp. 251–261. [Google Scholar]
- Kabil, O.; Banerjee, R. Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 2010, 285, 21903–21907. [Google Scholar] [CrossRef]
- Mustafa, A.K.; Gadalla, M.M.; Snyder, S.H. Signaling by gastransmitters. Sci. Signal. 2009, 2. [Google Scholar] [CrossRef]
- Kimura, H.; Shibuya, N.; Kimura, Y. Hydrogen sulfide is a signaling molecule and a cytoprotectant. Antioxid. Redox Sign. 2012, 17, 45–57. [Google Scholar] [CrossRef]
- Takahashi, H.; Sekimoto, M.; Tanaka, M.; Tanaka, A.; Igarashi, J.; Shimizu, T. Hydrogen sulfide stimulates the catalytic activity of a heme-regulated phosphodiesterase from Escherichia coli (Ec DOS). J. Inorg. Biochem. 2012, 109, 66–71. [Google Scholar] [CrossRef]
- Du, Y.; Liu, G.; Yan, Y.; Huang, D.; Luo, W.; Martinkova, M.; Man, P.; Shimizu, T. Conversion of a heme-based oxygen sensor to a heme oxygenase by hydrogen sulfide: Effects of mutatations in the heme distal side of a heme-based oxygen sensor phospodiesterase (Ec DOS). BioMetals 2013, in press. [Google Scholar]
- Watanabe, M.; Kurokawa, H.; Yoshimura-Suzuki, T.; Sagami, I.; Shimizu, T. Critical roles of Asp40 at the haem proximal side of haem-regulated phosphodiesterase from Escherichia coli in redox potential, auto-oxidation and catalytic control. Eur. J. Biochem. 2004, 271, 3937–3942. [Google Scholar] [CrossRef]
- Ito, S.; Igarashi, J.; Shimizu, T. The FG loop of a heme-based gas sensor enzyme, Ec DOS, functions in heme binding, autoxidation and catalysis. J. Inorg. Biochem. 2009, 103, 1380–1385. [Google Scholar] [CrossRef]
- Hao, B.; Isaza, C.; Arndt, J.; Soltis, M.; Chan, M.K. Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 2002, 41, 12952–12958. [Google Scholar] [CrossRef]
- Lechauve, C.; Bouzhi-Sima, L.; Yamashita, T.; Marden, M.C.; Vos, M.H.; Liebl, U.; Kiger, L. Heme ligand binding properties and intradimer interactions in the full-length sensor protein Dos from Escherichia coli and its isolated heme domain. J. Biol. Chem. 2009, 284, 36146–36159. [Google Scholar]
- Miksanova, M.; Igarashi, J.; Minami, M.; Sagami, I.; Yamauchi, S.; Kurokawa, H.; Shimizu, T. Characterization of heme-regulated eIF2α kinase: Roles of the N-terminal domain in the oligomeric state, heme binding, catalysis and inhibition. Biochemistry 2006, 45, 9894–9905. [Google Scholar]
- Taguchi, S.; Matsui, T.; Igarashi, J.; Sasakura, Y.; Araki, Y.; Ito, O.; Sugiyama, S.; Sagami, I.; Shimizu, T. Binding of oxygen and carbon monoxide to a heme-regulated phosphodiesterase from Escherichia coli: Kinetics and infrared spectra of the full-length wild-type enzyme, isolated PAS domain, and Met95 mutants. J. Biol. Chem. 2004, 279, 3340–3347. [Google Scholar]
- Kitanishi, K.; Kobayashi, K.; Uchida, T.; Ishimori, K.; Igarashi, J.; Shimizu, T. Identification and functional and spectral characterization of a globin-coupled histidine kinase from Anaeromyxobacter sp. Fw109–5. J. Biol. Chem. 2011, 286, 35522–35534. [Google Scholar]
- Nakajima, N.; Kitanishi, K.; Kobayashi, K.; Kobayashi, N.; Igarashi, J.; Shimizu, T. Leu65 in the heme distal side is critical for the stability of the Fe(II)-O2 complex of YddV, a globin-coupled oxygen sensor diguanylate cyclase. J. Inorg. Biochem. 2012, 108, 163–170. [Google Scholar] [CrossRef]
- Yokota, N.; Araki, Y.; Kurokawa, H.; Ito, O.; Igarashi, J.; Shimizu, T. Critical roles of Leu99 and Leu115 at the heme distal side in auto-oxidation and the redox potential of a heme-regulated phosphodiesterase from Escherichia coli. FEBS J. 2006, 273, 1210–1223. [Google Scholar] [CrossRef]
- Hirata, S.; Matsui, T.; Sasakura, Y.; Sugiyama, S.; Yoshimura, T.; Sagami, I.; Shimizu, T. Characterization of Met95 mutants of a heme-regulated phosphodiesterase from Escherichia coli: Optical absorption, magnetic circular dichroism, circular dichroism, and redox potentials. Eur. J. Biochem. 2003, 270, 4771–4779. [Google Scholar] [CrossRef]
- Gonzalez, G.; Dioum, E.M.; Bertolucci, C.M.; Tomita, T.; Ikeda-Saito, M.; Cheesman, M.R.; Watmough, N.J.; Gilles-Gonzalez, M.A. Nature of the displaceable heme-axial residue in the EcDOS protein, a heme-based sensor from Escherichia coli. Biochemistry 2002, 41, 8414–8421. [Google Scholar] [CrossRef]
- Ito, S. Molecular Mechanism of Oxygen Recognition and Intramolecular Signal Transduction of Heme-Regulated Oxygen-Sensor Enzyme, Ec DOS. Master Thesis, Tohoku University, Sendai, Japan, March 2009. [Google Scholar]
- Sousa, E.H.S.; Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.A. A memory of oxygen binding explains the dose response of the heme-based sensor FixL. Biochemistry 2007, 46, 6249–6257. [Google Scholar]
- Sousa, E.H.S.; Tuckerman, J.R.; Condim, A.C.S.; Gonzalez, G.; Gilles-Gonzalez, M.A. Signal transduction and phosphoryl transfer by a FixL hybrid kinase with low oxygen affinity: Importance of the vicinal PAS domain and receiver aspartate. Biochemistry 2013, 52, 456–465. [Google Scholar]
- Springer, B.A.; Sligar, S.G.; Olson, J.S.; Phillips, G.N., Jr. Mechanisms of ligand recognition in myoglobin. Chem. Rev. 1994, 94, 699–714. [Google Scholar]
- Shikama, K. The molecular mechanism of autooxidation for myoglobin and hemoglobin: A venerable puzzle. Chem. Rev. 1998, 98, 1357–1374. [Google Scholar] [CrossRef]
- Bidwai, A.K.; Ok, E.Y.; Erman, J.E. pH Dependence of cyanide binding to the ferric heme domain of the direct oxygen sensor from Escherichia coli and the effect of alkaline denaturation. Biochemistry 2008, 47, 10458–10470. [Google Scholar] [CrossRef]
- Watanabe, M.; Matsui, T.; Sasakura, Y.; Sagami, I.; Shimizu, T. Unusual cyanide bindings to a heme-regulated phosphodiesterase from Escherichia coli: Effect of Met95 mutations. Biochem. Biophys. Res. Commun. 2002, 299, 169–172. [Google Scholar]
- Sato, A.; Sasakura, Y.; Sugiyama, S.; Sagami, I.; Shimizu, T.; Mizutani, Y.; Kitagawa, T. Stationary and time-resolved resonance Raman spectra of His77 and Met95 mutants of the isolated heme domain of a direct oxygen sensor from E. coli. J. Biol. Chem. 2002, 277, 32650–32658. [Google Scholar]
- Tomita, T.; Gonzalez, G.; Chang, A.L.; Ikeda-Saito, M.; Gilles-Gonzalez, M.A. A comparative resonance Raman analysis of heme-binding PAS domains: Heme iron coordination structure of the Bj FixL, Ax PDEA1, Ec DOS, and Mt Dos proteins. Biochemistry 2002, 41, 4819–4826. [Google Scholar]
- El-Mashtoly, S.F.; Nakashima, S.; Tanaka, A.; Shimizu, T.; Kitagawa, T. Roles of Arg97 and Phe113 in regulation of distal ligand binding to heme in the sensor domain of Ec DOS protein: Resonance Raman and mutation study. J. Biol. Chem. 2008, 283, 19000–19010. [Google Scholar]
- El-Mashtoly, S.F.; Takahashi, H.; Shimizu, T.; Kitagawa, T. Ultraviolet resonance Raman evidence for utilization of the heme 6-propionate hydrogen-bond network in signal transmission from heme to protein in Ec DOS protein. J. Am. Chem. Soc. 2007, 129, 3556–3563. [Google Scholar]
- El-Mashtoly, S.F.; Takahashi, H.; Kurokawa, H.; Sato, A.; Shimizu, T.; Kitagawa, T. Resonance Raman investigation of redox-induced structural changes of protein and heme in the sensor domain of Ec DOS protein. J. Raman Spectrosc. 2008, 39, 1614–1626. [Google Scholar] [CrossRef]
- El-Mashtoly, S.F.; Kudo, M.; Nakashima, S.; Shimizu, T.; Kitagawa, T. Structural dynamics of Ec DOS heme domain revealed by time-resolved ultraviolet resonance Raman spectroscopy. J. Phys. Chem. Lett. 2011, 2, 2212–2217. [Google Scholar] [CrossRef]
- Kobayashi, K.; Tagawa, S.; Daff, S.; Sagami, I.; Shimizu, T. Rapid calmodulin-dependent interdomain electron transfer in neuronal nitric-oxide synthase measured by pulse radiolysis. J. Biol. Chem. 2001, 276, 39864–39871. [Google Scholar]
- Kobayashi, K.; Tanaka, A.; Takahashi, H.; Igarashi, J.; Ishitsuka, Y.; Yokota, N.; Shimizu, T. Catalysis and oxygen binding of Ec DOS, a heme-based oxygen-sensor enzyme from Escherichia coli. J. Biochem. 2010, 148, 693–703. [Google Scholar]
- Yamashita, T.; Bouzhir-Sima, L.; Lambry, J.-C.; Liebl, U.; Vos, M.H. Ligand dynamics and early signaling events in the heme domain of the sensor protein Dos from Escherichia coli. J. Biol. Chem. 2008, 283, 2344–2352. [Google Scholar]
- Liebl, U.; Bouzhir-Sima, L.; Kiger, L.; Marden, M.C.; Lambry, J.-C.; Négrerie, M.; Vos, M.H. Ligand binding dynamics to the heme domain of the oxygen sensor Dos from Escherichia coli. Biochemistry 2003, 42, 6527–6535. [Google Scholar]
- Liebl, U.; Bouzhir-Sima, L.; Négrerie, M.; Martin, J.-L.; Vos, M.H. Ultrafast ligand rebinding in the heme domain of the oxygen sensors FixL and Dos: General regulatory implications for heme-based sensors. Proc. Nat. Acad. Sci. USA 2002, 99, 12771–12776. [Google Scholar]
- Ito, S.; Araki, Y.; Tanaka, A.; Igarashi, J.; Wada, T.; Shimizu, T. Role of Phe113 at the distal side of the heme domain of an oxygen-sensor (Ec DOS) in the characterization of the heme environment. J. Inorg. Biochem. 2009, 103, 989–996. [Google Scholar] [CrossRef]
- Hirata, S.; Kurakawa, H.; Sagami, I.; Shimizu, T. Fluorescence spectral studies of Trp53Phe and Trp110Ile mutants of a heme-regulated phosphodiesterase from Escherichia coli. Chem. Lett. 2005, 34, 752–753. [Google Scholar]
- Yoshimura-Suzuki, Y.; Sagami, I.; Yokota, N.; Kurokawa, H.; Shimizu, T. DOSEc, a heme-regulated phosphodiesterase, plays an important role in the regulation of the cyclic AMP level in Escherichia coli. J. Bacteriol. 2005, 187, 6678–6682. [Google Scholar]
- Yoshimura-Suzuki, K. Biochemical characterization and physiological role of a heme-regulated phosphodiesterase from Escherichia coli (Ec DOS). Ph.D. Thesis, Tohoku University, Sendai, Japan, March 2004. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shimizu, T. The Heme-Based Oxygen-Sensor Phosphodiesterase Ec DOS (DosP): Structure-Function Relationships. Biosensors 2013, 3, 211-237. https://doi.org/10.3390/bios3020211
Shimizu T. The Heme-Based Oxygen-Sensor Phosphodiesterase Ec DOS (DosP): Structure-Function Relationships. Biosensors. 2013; 3(2):211-237. https://doi.org/10.3390/bios3020211
Chicago/Turabian StyleShimizu, Toru. 2013. "The Heme-Based Oxygen-Sensor Phosphodiesterase Ec DOS (DosP): Structure-Function Relationships" Biosensors 3, no. 2: 211-237. https://doi.org/10.3390/bios3020211
APA StyleShimizu, T. (2013). The Heme-Based Oxygen-Sensor Phosphodiesterase Ec DOS (DosP): Structure-Function Relationships. Biosensors, 3(2), 211-237. https://doi.org/10.3390/bios3020211




