Changes in Cis-regulatory Elements during Morphological Evolution
Abstract
:1. Introduction
2. Examples of Changes in Cis-regulatory Elements during Morphological Evolution
2.1. Evolutionary Change Recognized as Change in a Cis-regulatory Element of a Developmental Gene
2.1.1. Stickleback Pelvic Fin Loss

2.1.2. Forelimb Length in Bat and Mouse
2.1.3. Vertebral Formulae of Mouse and Chicken
2.1.4. Loss of Vibrissae and Penile Spines in Human
2.3. Evolutionary Change Understood as Acquisition, Loss, or Changed Efficiency of Defined Functional Motifs within Cis-regulatory Elements
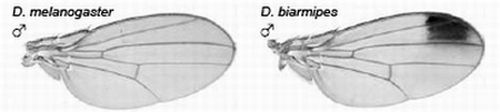
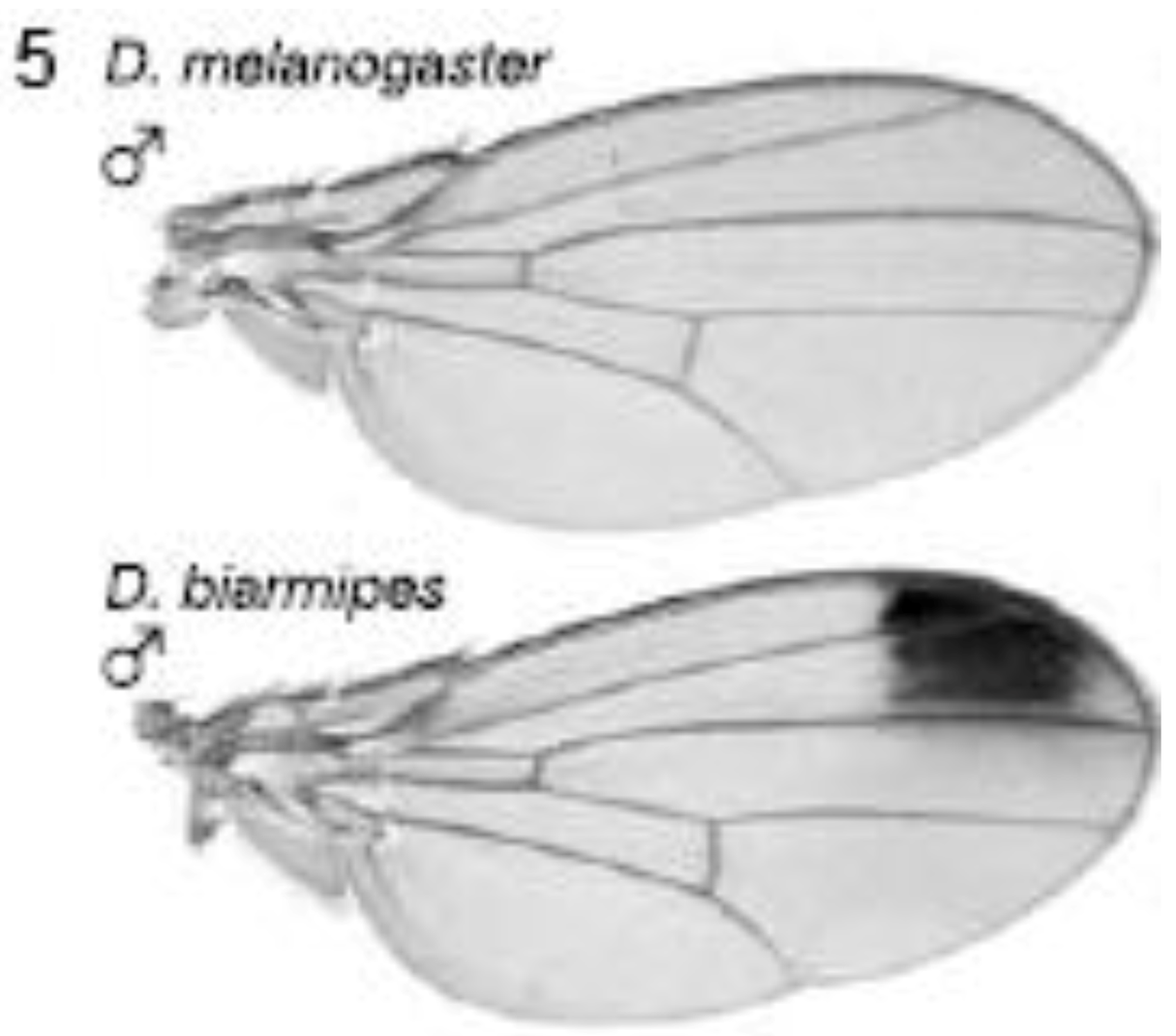
2.3.1. Male-specific Wing Spots in Drosophila biarmipes
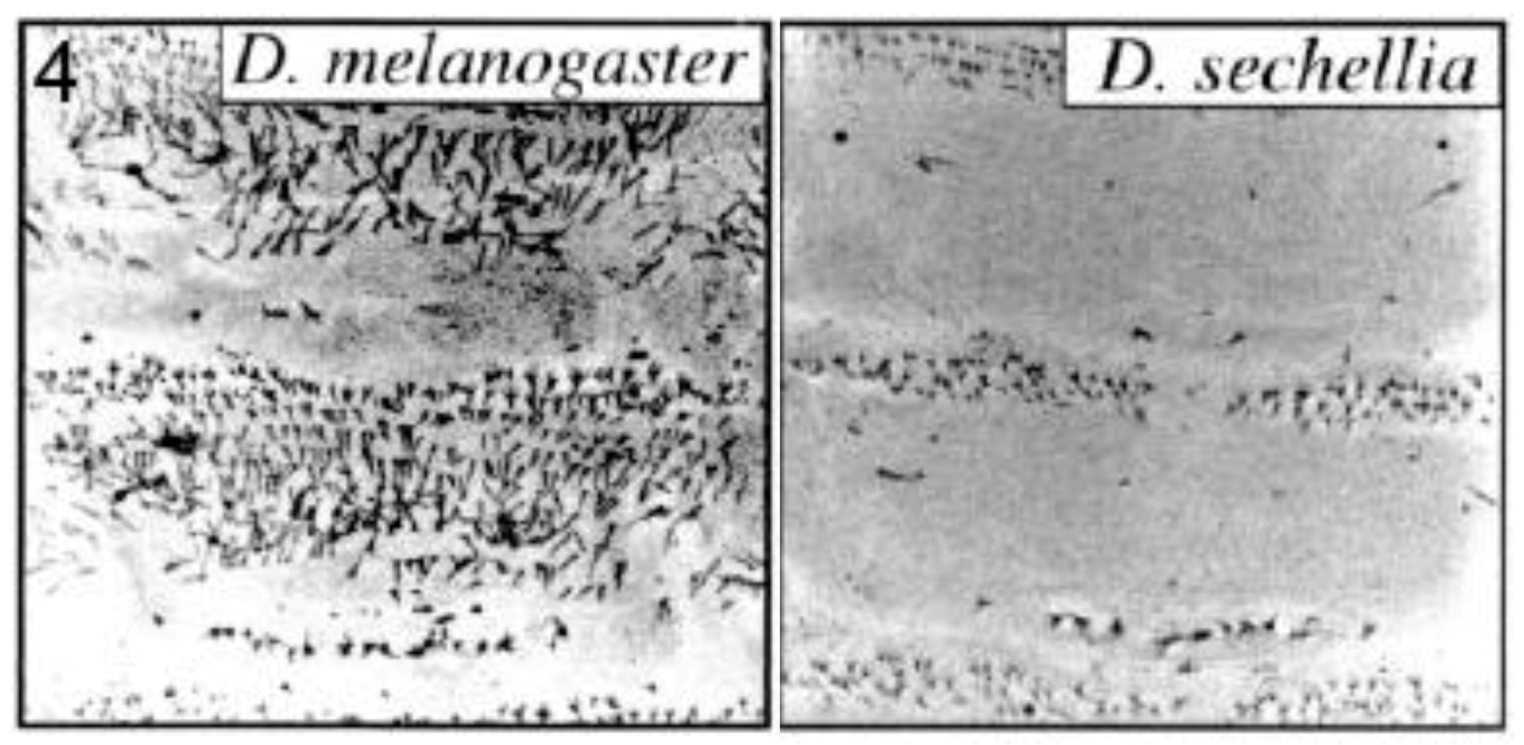
2.3.2. Male-specific Abdominal Pigmentation Differences between Drosophila melanogaster and D. willistoni

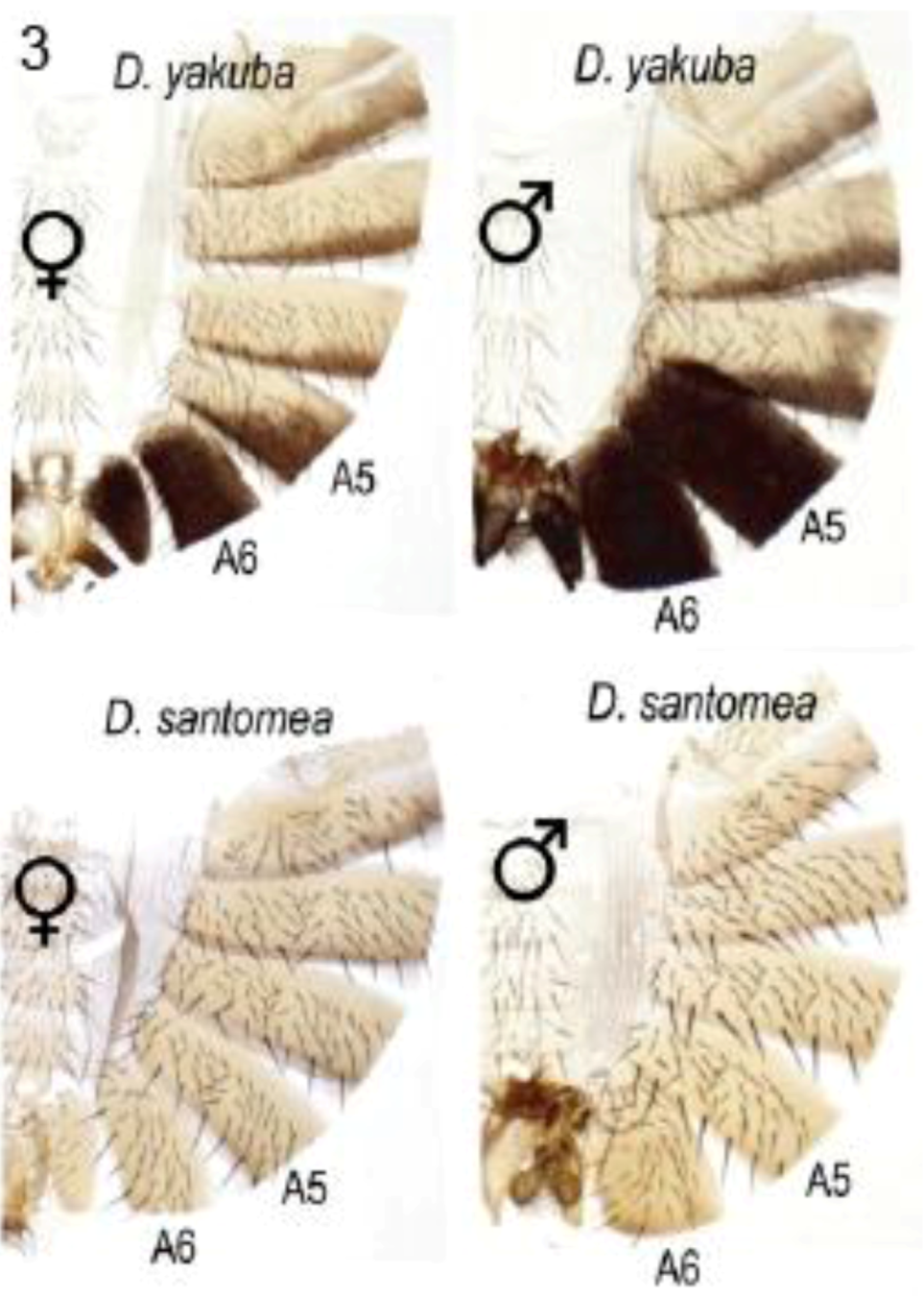
2.3.3. Abdominal Pigmentation Differences between Drosophila melanogaster and D. kikkawai Species
2.4. Phylogenetic Modification/Appearance/Loss of a Cis-regulatory Element which is of Suggestive but Unproven Function
2.4.1. Human Specific Mutations within a Limb Cis-regulatory Element
2.4.2. Sox Response Elements in Fezf2, a Determinant of Corticospinal Neuron Identity
2.4.3. Retinoic Acid Response Elements in Two Cis-regulatory Elements of the Vertebrate Homeobox Gene, Cdx1
| Fish | Amphibian | Lizard | Bird | Marsupial | Eutherian | References | |
|---|---|---|---|---|---|---|---|
| Cdx1 upstream | ✓ | [47], [38] | |||||
| Cdx1 intron | ✓ | ✓ | ✓ | ✓ | [38] | ||
| Hoxa1 (3’ DR5) | ?* |  |  |  | ✓ | [45] | |
| Hoxa4 (5’ DR5) | ✓ |  |  |  |  | ✓ | [48] |
| Hoxb1 (3’ DR2) | ✓ |  |  | ✓ | [49] | ||
| Hoxb1 (3’ DR5) | ✓** |  | ✓ |  | ✓ | [50], [45] | |
| Hoxb1 (5’ DR2) | ✓ | ✓ | [51] | ||||
| Hoxb4 (3’ DR5) | ✓ |  |  |  | ✓ | [52] | |
| Hoxb5 (3’ DR5) |  |  |  | ✓ | [53] | ||
| Hoxd4 (5’ DR5) | ✓*** |  |  |  |  | ✓ | [46] |
| Hoxd4 (3’ DR5) | ✓*** |  |  |  |  | ✓ | [46] |
2.4.4. Human-specific Loss of a CRE in the GADD45G Tumour Suppressor Gene
3. Discussion
3.1. Modification of Cis-regulatory Elements Revealed as a Mechanism Used in Morphological Evolution
3.2. A Problem in Comparing Cis-regulatory Elements of Distantly Related Species
3.3. Evolutionarily Origins of Cis-regulatory Elements Identified in Database Searches
3.4. Problems in Elucidating the Evolutionary Basis of Widely Divergent Morphologies
4. Conclusions
References and Notes
- Carroll, S.B.; Grenier, J.K.; Weatherbee, S.D. From DNA to diversity: Molecular Genetics and the Evolution of Animal Design, 2nd; Sean, B.C., Jennifer, K.G., Eds.; Blackwell Science Press: Madison, WI, USA, 2005. [Google Scholar]
- Carroll, S.B. Evolution at two levels: On genes and form. PloS Biol. 2005. [Google Scholar] [CrossRef] [Green Version]
- Prud'homme, B.; Gompel, N.; Carroll, S.B. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 8605–8612. [Google Scholar]
- Wray, G.A.; Hahn, M.W.; Abouheif, E.; Balhoff, J.P.; Pizer, M.; Rockman, M.V.; Romano, L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. [Google Scholar] [CrossRef]
- Galant, R.; Carroll, S.B. Evolution of a transcriptional repression domain in an insect hox protein. Nature 2002, 415, 910–913. [Google Scholar] [CrossRef]
- Ronshaugen, M.; McGinnis, N.; McGinnis, W. Hox protein mutation and macroevolution of the insect body plan. Nature 2002, 415, 914–917. [Google Scholar] [CrossRef]
- Shiga, Y.; Yasumoto, R.; Yamagata, H.; Hayashi, S. Evolving role of antennapedia protein in arthropod limb patterning. Development 2002, 129, 3555–3561. [Google Scholar]
- Lynch, V.J.; Wagner, G.P. Resurrecting the role of transcription factor change in developmental evolution. Evolution 2008, 62, 2131–2154. [Google Scholar] [CrossRef]
- Monteiro, A.; Podlaha, O. Wings, horns, and butterfly eyespots: How do complex traits evolve? Plos Biol. 2009. [Google Scholar] [CrossRef]
- Hoekstra, H.E.; Coyne, J.A. The locus of evolution: Evo devo and the genetics of adaptation. Evolution 2007, 61, 995–1016. [Google Scholar] [CrossRef]
- Pennisi, E. Deciphering the genetics of evolution. Science 2008, 321, 760–763. [Google Scholar] [CrossRef]
- Pick, L.; Heffer, A. Hox gene evolution: Multiple mechanisms contributing to evolutionary novelties. Ann. NY Acad. Sci. 2012, 1256, 15–32. [Google Scholar]
- Chan, Y.F.; Marks, M.E.; Jones, F.C.; Villarreal, G., Jr.; Shapiro, M.D.; Brady, S.D.; Southwick, A.M.; Absher, D.M.; Grimwood, J.; Schmutz, J.; et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a pitx1 enhancer. Science 2010, 327, 302–305. [Google Scholar]
- Shapiro, M.D.; Bell, M.A.; Kingsley, D.M. Parallel genetic origins of pelvic reduction in vertebrates. Proc. Natl. Acad. Sci. USA 2006, 103, 13753–13758. [Google Scholar]
- Cretekos, C.J.; Wang, Y.; Green, E.D.; Martin, J.F.; Rasweiler, J.J.T.; Behringer, R.R. Regulatory divergence modifies limb length between mammals. Genes Dev. 2008, 22, 141–151. [Google Scholar] [CrossRef]
- Gaunt, S.J. Conservation in the hox code during morphological evolution. Int. J. Dev. Biol. 1994, 38, 549–552. [Google Scholar]
- Burke, A.C.; Nelson, C.E.; Morgan, B.A.; Tabin, C. Hox genes and the evolution of vertebrate axial morphology. Development 1995, 121, 333–346. [Google Scholar]
- Gaunt, S.J.; Dean, W.; Sang, H.; Burton, R.D. Evidence that hoxa expression domains are evolutionarily transposed in spinal ganglia, and are established by forward spreading in paraxial mesoderm. Mech. Develop. 1999, 82, 109–118. [Google Scholar] [CrossRef]
- Belting, H.G.; Shashikant, C.S.; Ruddle, F.H. Modification of expression and cis-regulation of hoxc8 in the evolution of diverged axial morphology. Proc. Natl. Acad. Sci. USA 1998, 95, 2355–2360. [Google Scholar]
- Shashikant, C.S.; Bolanowsky, S.A.; Anand, S.; Anderson, S.M. Comparison of diverged hoxc8 early enhancer activities reveals modification of regulatory interactions at conserved cis-acting elements. J. Exp. Zool. B. Mol. Dev. Evol. 2007, 308, 242–249. [Google Scholar]
- Shashikant, C.S.; Kim, C.B.; Borbely, M.A.; Wang, W.C.; Ruddle, F.H. Comparative studies on mammalian hoxc8 early enhancer sequence reveal a baleen whale-specific deletion of a cis-acting element. Proc. Natl. Acad. Sci. USA 1998, 95, 15446–15451. [Google Scholar]
- Anand, S.; Wang, W.C.; Powell, D.R.; Bolanowski, S.A.; Zhang, J.; Ledje, C.; Pawashe, A.B.; Amemiya, C.T.; Shashikant, C.S. Divergence of hoxc8 early enhancer parallels diverged axial morphologies between mammals and fishes. Proc. Natl. Acad. Sci. USA 2003, 100, 15666–15669. [Google Scholar]
- McLean, C.Y.; Reno, P.L.; Pollen, A.A.; Bassan, A.I.; Capellini, T.D.; Guenther, C.; Indjeian, V.B.; Lim, X.; Menke, D.B.; Schaar, B.T.; et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 2011, 471, 216–219. [Google Scholar] [CrossRef]
- Rebeiz, M.; Pool, J.E.; Kassner, V.A.; Aquadro, C.F.; Carroll, S.B. Stepwise modification of a modular enhancer underlies adaptation in a drosophila population. Science 2009, 326, 1663–1667. [Google Scholar] [CrossRef]
- McGregor, A.P.; Orgogozo, V.; Delon, I.; Zanet, J.; Srinivasan, D.G.; Payre, F.; Stern, D.L. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 2007, 448, 587–590. [Google Scholar]
- Williams, T.M.; Selegue, J.E.; Werner, T.; Gompel, N.; Kopp, A.; Carroll, S.B. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in drosophila. Cell 2008, 134, 610–623. [Google Scholar] [CrossRef]
- Prabhakar, S.; Visel, A.; Akiyama, J.A.; Shoukry, M.; Lewis, K.D.; Holt, A.; Plajzer-Frick, I.; Morrison, H.; Fitzpatrick, D.R.; Afzal, V.; et al. Human-specific gain of function in a developmental enhancer. Science 2008, 321, 1346–1350. [Google Scholar]
- Frankel, N.; Erezyilmaz, D.F.; McGregor, A.P.; Wang, S.; Payre, F.; Stern, D.L. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 2011, 474, 598–603. [Google Scholar]
- Jeong, S.; Rokas, A.; Carroll, S.B. Regulation of body pigmentation by the abdominal-b hox protein and its gain and loss in drosophila evolution. Cell 2006, 125, 1387–1399. [Google Scholar] [CrossRef]
- Jeong, S.; Rebeiz, M.; Andolfatto, P.; Werner, T.; True, J.; Carroll, S.B. The evolution of gene regulation underlies a morphological difference between two drosophila sister species. Cell 2008, 132, 783–793. [Google Scholar] [CrossRef]
- Sucena, E.; Stern, D.L. Divergence of larval morphology between drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. USA 2000, 97, 4530–4534. [Google Scholar] [CrossRef]
- Gompel, N.; Prud'homme, B.; Wittkopp, P.J.; Kassner, V.A.; Carroll, S.B. Chance caught on the wing: Cis-regulatory evolution and the origin of pigment patterns in drosophila. Nature 2005, 433, 481–487. [Google Scholar]
- Sumiyama, K.; Saitou, N. Loss-of-function mutation in a repressor module of human-specifically activated enhancer hacns1. Mol. Biol. Evol. 2011, 28, 3005–3007. [Google Scholar] [CrossRef]
- Shim, S.; Kwan, K.Y.; Li, M.; Lefebvre, V.; Sestan, N. Cis-regulatory control of corticospinal system development and evolution. Nature 2012, 486, 74–79. [Google Scholar]
- Gaunt, S.J.; Drage, D.; Cockley, A. Vertebrate caudal gene expression gradients investigated by use of chick cdx-a/lacz and mouse cdx-1/lacz reporters in transgenic mouse embryos: Evidence for an intron enhancer. Mech. Dev. 2003, 120, 573–586. [Google Scholar] [CrossRef]
- Houle, M.; Sylvestre, J.R.; Lohnes, D. Retinoic acid regulates a subset of cdx1 function in vivo. Development 2003, 130, 6555–6567. [Google Scholar] [CrossRef]
- Marletaz, F.; Holland, L.Z.; Laudet, V.; Schubert, M. Retinoic acid signaling and the evolution of chordates. Int. J.Biol. Sci. 2006, 2, 38–47. [Google Scholar]
- Gaunt, S.J.; Paul, Y.L. Origins of cdx1 regulatory elements suggest roles in vertebrate evolution. Int. J. Dev. Biol. 2011, 55, 93–98. [Google Scholar] [CrossRef]
- Gaunt, S.J.; Drage, D.; Trubshaw, R.C. Increased cdx protein dose effects upon axial patterning in transgenic lines of mice. Development 2008, 135, 2511–2520. [Google Scholar] [CrossRef]
- Evans, F.G. The morphology and functional evolution of the atlas-axis complex from fish to mammals. Ann. N. Y. Acad. Sci. 1939, 39, 29–104. [Google Scholar] [CrossRef]
- Sumida, S.S.; Lombard, R.E. The atlas-axis complex in the late paleozoic genus diadectes and the characteristics of the atlas-axis complex across the amphibian to amniote transition. J. Paleontol. 1991, 65, 973–983. [Google Scholar]
- Vandepoele, K.; De Vos, W.; Taylor, J.S.; Meyer, A.; Van de Peer, Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. USA 2004, 101, 1638–1643. [Google Scholar]
- McClintock, J.M.; Carlson, R.; Mann, D.M.; Prince, V.E. Consequences of hox gene duplication in the vertebrates: An investigation of the zebrafish hox paralogue group 1 genes. Development 2001, 128, 2471–2484. [Google Scholar]
- McClintock, J.M.; Kheirbek, M.A.; Prince, V.E. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development 2002, 129, 2339–2354. [Google Scholar]
- Ishioka, A.; Jindo, T.; Kawanabe, T.; Hatta, K.; Parvin, M.S.; Nikaido, M.; Kuroyanagi, Y.; Takeda, H.; Yamasu, K. Retinoic acid-dependent establishment of positional information in the hindbrain was conserved during vertebrate evolution. Dev. Biol. 2011, 350, 154–168. [Google Scholar] [CrossRef]
- Nolte, C.; Amores, A.; Nagy Kovacs, E.; Postlethwait, J.; Featherstone, M. The role of a retinoic acid response element in establishing the anterior neural expression border of hoxd4 transgenes. Mech. Dev. 2003, 120, 325–335. [Google Scholar] [CrossRef]
- Houle, M.; Prinos, P.; Iulianella, A.; Bouchard, N.; Lohnes, D. Retinoic acid regulation of cdx1: An indirect mechanism for retinoids and vertebral specification. Mol. Cell Biol. 2000, 20, 6579–6586. [Google Scholar] [CrossRef]
- Packer, A.I.; Crotty, D.A.; Elwell, V.A.; Wolgemuth, D.J. Expression of the murine hoxa4 gene requires both autoregulation and a conserved retinoic acid response element. Development 1998, 125, 1991–1998. [Google Scholar]
- Marshall, H.; Studer, M.; Popperl, H.; Aparicio, S.; Kuroiwa, A.; Brenner, S.; Krumlauf, R. A conserved retinoic acid response element required for early expression of the homeobox gene hoxb-1. Nature 1994, 370, 567–571. [Google Scholar]
- Langston, A.W.; Thompson, J.R.; Gudas, L.J. Retinoic acid-responsive enhancers located 3' of the hox a and hox b homeobox gene clusters. Functional analysis. J. Biol. Chem. 1997, 272, 2167–2175. [Google Scholar]
- Studer, M.; Popperl, H.; Marshall, H.; Kuroiwa, A.; Krumlauf, R. Role of a conserved retinoic acid response element in rhombomere restriction of hoxb-1. Science 1994, 265, 1728–1732. [Google Scholar]
- Gould, A.; Itasaki, N.; Krumlauf, R. Initiation of rhombomeric hoxb4 expression requires induction by somites and a retinoid pathway. Neuron 1998, 21, 39–51. [Google Scholar] [CrossRef]
- Oosterveen, T.; Niederreither, K.; Dolle, P.; Chambon, P.; Meijlink, F.; Deschamps, J. Retinoids regulate the anterior expression boundaries of 5' hoxb genes in posterior hindbrain. EMBO. J. 2003, 22, 262–269. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar]
- Simpson, P. Evolution of development in closely related species of flies and worms. Nature Rev. Genet. 2002, 3, 907–917. [Google Scholar] [CrossRef]
- Morrison, A.; Chaudhuri, C.; Ariza-McNaughton, L.; Muchamore, I.; Kuroiwa, A.; Krumlauf, R. Comparative analysis of chicken hoxb-4 regulation in transgenic mice. Mech. Dev. 1995, 53, 47–59. [Google Scholar] [CrossRef]
- Whiting, J.; Marshall, H.; Cook, M.; Krumlauf, R.; Rigby, P.W.; Stott, D.; Allemann, R.K. Multiple spatially specific enhancers are required to reconstruct the pattern of hox-2.6 gene expression. Genes Dev. 1991, 5, 2048–2059. [Google Scholar] [CrossRef]
- Gilbert, W.; Marchionni, M.; McKnight, G. On the antiquity of introns. Cell 1986, 46, 151–153. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life, 5th; Murray, J., Ed.; The University of Oxford: London, UK, 1859. [Google Scholar]
- Subramanian, V.; Meyer, B.I.; Gruss, P. Disruption of the murine homeobox gene cdx1 affects axial skeletal identities by altering the mesodermal expression domains of hox genes. Cell 1995, 83, 641–653. [Google Scholar] [CrossRef]
Supplementary Materials

© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gaunt, S.J.; Paul, Y.-L. Changes in Cis-regulatory Elements during Morphological Evolution. Biology 2012, 1, 557-574. https://doi.org/10.3390/biology1030557
Gaunt SJ, Paul Y-L. Changes in Cis-regulatory Elements during Morphological Evolution. Biology. 2012; 1(3):557-574. https://doi.org/10.3390/biology1030557
Chicago/Turabian StyleGaunt, Stephen J., and Yu-Lee Paul. 2012. "Changes in Cis-regulatory Elements during Morphological Evolution" Biology 1, no. 3: 557-574. https://doi.org/10.3390/biology1030557