In Silico Research of New Therapeutics Rotenoids Derivatives against Leishmania amazonensis Infection
Abstract
:Simple Summary
Abstract
1. Introduction
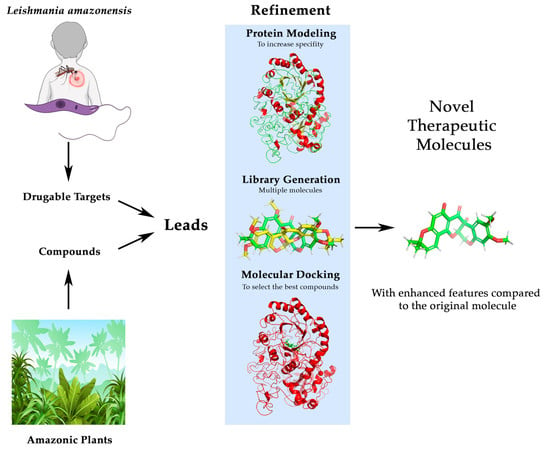
2. Materials and Methods
2.1. Bibliographical Review and Identification of Leads
2.2. Lead Compound Improvement and Enhancement
3. Results
3.1. Selected Target and Query Phytocompound
3.2. Pharmacological Optimization of Deguelin
3.2.1. Three-Dimensional Structures
3.2.2. Assessment of ORF Sequences for L. amazonensis ODC
3.2.3. Identification and Assessment of ODC and NUO Binding to Compounds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torres-guerrero, E.; Quintanilla-cedillo, M.R.; Ruiz-esmenjaud, J.; Arenas, R. Leishmaniasis: A Review. F1000 Rev. 2017, 6, 750. [Google Scholar] [CrossRef]
- Roberts, L.J.; Handman, E.; Foote, S.J. Science, Medicine, and the Future: Leishmaniasis. BMJ 2000, 321, 801–804. [Google Scholar] [CrossRef] [PubMed]
- WHO Leishmaniasis (Updated 20 May 2021). Available online: https://www.who.int/es/news-room/fact-sheets/detail/leishmaniasis (accessed on 3 November 2021).
- WHO Leishmaniasis-Status of Endemicity of Cutaneous Leishmaniasis: 2020. Available online: https://apps.who.int/neglected_diseases/ntddata/leishmaniasis/leishmaniasis.html (accessed on 3 November 2021).
- Akhoundi, M.; Kuhls, K.; Cannet, A.; Votýpka, J.; Marty, P.; Delaunay, P.; Sereno, D. A Historical Overview of the Classification, Evolution, and Dispersion of Leishmania Parasites and Sandflies. PLoS Negl. Trop. Dis. 2016, 10, e0004349. [Google Scholar] [CrossRef]
- Christensen, S.M.; Belew, A.T.; El-Sayed, N.M.; Tafuri, W.L.; Silveira, F.T.; Mosser, D.M. Host and Parasite Responses in Human Diffuse Cutaneous Leishmaniasis Caused by L. Amazonensis. PLoS Negl. Trop. Dis. 2019, 13, e0007152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, J.I.; Hong, A.; Zampieri, R.A.; Floeter-Winter, L.M.; Laranjeira-Silva, M.F. In Vivo Infection with Leishmania Amazonensis to Evaluate Parasite Virulence in Mice. J. Vis. Exp. 2020, 2020, e60617. [Google Scholar] [CrossRef] [PubMed]
- Camara Coelho, L.I.; Paes, M.; Guerra, J.A.; Barbosa, M.d.G.; Coelho, C.; Lima, B.; Brito, M.E.; Brandão Filho, S.P. Characterization of Leishmania Spp. Causing Cutaneous Leishmaniasis in Manaus, Amazonas, Brazil. Parasitol. Res. 2011, 108, 671–677. [Google Scholar] [CrossRef] [Green Version]
- Valdivia, H.O.; Almeida, L.V.; Roatt, B.M.; Reis-Cunha, J.L.; Pereira, A.A.S.; Gontijo, C.; Fujiwara, R.T.; Reis, A.B.; Sanders, M.J.; Cotton, J.A.; et al. Comparative Genomics of Canine-Isolated Leishmania (Leishmania) Amazonensis from an Endemic Focus of Visceral Leishmaniasis in Governador Valadares, Southeastern Brazil. Sci. Rep. 2017, 7, 40804. [Google Scholar] [CrossRef] [Green Version]
- Volpini, Â.C.; Passos, V.M.A.; Oliveira, G.C.; Romanha, A.J. PCR-RFLP to Identify Leishmania (Viannia) Braziliensis and L. (Leishmania) Amazonensis Causing American Cutaneous Leishmaniasis. Acta Trop. 2004, 90, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Barral, A.; Badaro, R.; Carvalho, E.M.; Barral-Netto, M.; de Jesus, A.R.; Johnson, W.D.; Momen, H.; Almeida, R.; McMahon-Pratt, D.; Pedral-Sampaio, D.; et al. Leishmaniasis in Bahia, Brazil: Evidence That Leishmania Amazonensis Produces a Wide Spectrum of Clinical Disease. Am. J. Trop. Med. Hyg. 1991, 44, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Ponte-Sucre, A.; Gamarro, F.; Dujardin, J.; Barrett, M.P.; Garcı, R.; Pountain, A.W.; Mwenechanya, R.; Papadopoulou, B. Drug Resistance and Treatment Failure in Leishmaniasis: A 21st Century Challenge. PLoS Negl. Trop. Dis. 2017, 11, 1–24. [Google Scholar] [CrossRef]
- Croft, S.L.; Sundar, S.; Fairlamb, A.H. Drug Resistance in Leishmaniasis. Clin. Microbiol. Rev. 2006, 19, 111–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hefnawy, A.; Berg, M.; Dujardin, J.; Muylder, G. De Exploiting Knowledge on Leishmania Drug Resistance to Support the Quest for New Drugs. Trends Parasitol. 2017, 33, 162–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapil, S.; Singh, P.K.; Silakari, O. An Update on Small Molecule Strategies Targeting Leishmaniasis. Eur. J. Med. Chem. 2018, 157, 339–367. [Google Scholar] [CrossRef]
- Stone, N.R.H.; Bicanic, T.; Salim, R.; Hope, W. Liposomal Amphotericin B (AmBisome ®): A Review of the Pharmacokinetics, Pharmacodynamics, Clinical Experience and Future Directions. Drugs 2017, 76, 485–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler-Moore, J.P.; Proffitt, R.T. Development, Characterization, Efficacy and Mode of Action of Ambisome, a Unilamellar Liposomal Formulation of Amphotericin B. J. Liposome Res. 1993, 3, 429–450. [Google Scholar] [CrossRef]
- Pinto-Martinez, A.K.; Rodriguez-Durán, J.; Serrano-Martin, X.; Hernandez-Rodriguez, V.; Benaim, G. Mechanism of Action of Miltefosine on Leishmania Donovani Involves the Impairment of Acidocalcisome Function and the Activation of the Sphingosine-Dependent Plasma Membrane Ca2+ Channel. Antimicrob. Agents Chemother. 2018, 62, e01614-17. [Google Scholar] [CrossRef] [Green Version]
- Mehta, R.; Champney, W.S. Neomycin and Paromomycin Inhibit 30S Ribosomal Subunit Assembly in Staphylococcus Aureus. Curr. Microbiol. 2003, 47, 237–243. [Google Scholar] [CrossRef]
- Maarouf, M.; de Kouchkovsky, Y.; Brown, S.; Petit, P.X.; Robert-Gero, M. In Vivo Interference of Paromomycin with Mitochondrial Activity of Leishmania. Exp. Cell Res. 1997, 232, 339–348. [Google Scholar] [CrossRef]
- Sands, M.; Kron, M.A.; Brown, R.B. Pentamidine: A Review. Rev. Infect. Dis. 1985, 7, 625–6344. [Google Scholar] [CrossRef]
- Rahman, I.U.; Afzal, A.; Iqbal, Z.; Ijaz, F.; Ali, N.; Shah, M.; Ullah, S.; Bussmann, R.W. Historical Perspectives of Ethnobotany. Clin. Dermatol. 2019, 37, 382–388. [Google Scholar] [CrossRef]
- Borris, R.P. Natural Products Research: Perspectives from a Major Pharmaceutical Company. J. Ethnopharmacol. 1996, 51, 29–38. [Google Scholar] [CrossRef]
- Heinrich, M.; Jäger, A.K. Ethnopharmacology; Heinrich, M., Jäger, A.K., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2015; ISBN 9781118930717. [Google Scholar]
- Kapetanovic, I.M. Computer-Aided Drug Discovery and Development (CADDD): In Silico-Chemico-Biological Approach. Chem. Biol. Interact. 2008, 171, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided-Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Kaur, E.A.; Khehra, B.S. Aproaches To Prediction Of Protein Structure: A Review. Int. Res. J. Eng. Technol. 2017, 4, 3564–3580. [Google Scholar]
- Palma, L.C.; Ferreira, L.F.G.R.; de Oliveira Almeida Petersen, A.L.; Dias, B.R.S.; de Menezes, J.P.B.; de Magalhães Moreira, D.R.; Hernandes, M.Z.; Veras, P.S.T. A Docking-Based Structural Analysis of Geldanamycin-Derived Inhibitor Binding to Human or Leishmania Hsp90. Sci. Rep. 2019, 9, 14756. [Google Scholar] [CrossRef]
- Morales-Jadán, D.; Blanco-Salas, J.; Ruiz-Téllez, T.; Centeno, F. Three Alkaloids from an Apocynaceae Species, Aspidosperma Spruceanum as Antileishmaniasis Agents by In Silico Demo-Case Studies. Plants 2020, 9, 983. [Google Scholar] [CrossRef] [PubMed]
- Gachet, M.S.; Lecaro, J.S.; Kaiser, M.; Brun, R.; Navarrete, H.; Muñoz, R.A.; Bauer, R.; Schühly, W. Assessment of Anti-Protozoal Activity of Plants Traditionally Used in Ecuador in the Treatment of Leishmaniasis. J. Ethnopharmacol. 2010, 128, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Luzuriaga Quichimbo, C.X. Estudio Etnobotánico En Comunidades Kichwas Amazónicas de Pastaza, Ecuador. Ph.D. Thesis, Universidad de Extremadura, Badajoz, Spain, 2017. [Google Scholar]
- Carvalho, B.M.; Rangel, E.F.; Ready, P.D.; Vale, M.M. Ecological Niche Modelling Predicts Southward Expansion of Lutzomyia (Nyssomyia) Flaviscutellata (Diptera: Psychodidae: Phlebotominae), Vector of Leishmania (Leishmania) Amazonensis in South America, under Climate Change. PLoS ONE 2015, 10, e0143282. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y. I-TASSER Server for Protein 3D Structure Prediction. BMC Bioinform. 2008, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Feig, M. Local Protein Structure Refinement via Molecular Dynamics Simulations with LocPREFMD. J. Chem. Inf. Model. 2016, 56, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the Estimation of the Absolute Quality of Individual Protein Structure Models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Schüller, A.; Hähnke, V.; Schneider, G. SmiLib v2. 0: A Java-Based Tool for Rapid Combinatorial Library Enumeration. QSAR Comb. Sci. 2007, 26, 407–410. [Google Scholar] [CrossRef]
- MOLINSPIRATION Collection of Substituents and Spacers Extracted from Bioactive Molecules. Available online: https://www.molinspiration.com/docu/fragments/index.html (accessed on 11 February 2019).
- Guedes, I.A.; Barreto, A.M.S.; Marinho, D.; Krempser, E.; Kuenemann, M.A.; Sperandio, O.; Dardenne, L.E.; Miteva, M.A. New Machine Learning and Physics-Based Scoring Functions for Drug Discovery. Sci. Rep. 2021, 11, 3198. [Google Scholar] [CrossRef] [PubMed]
- Santos, K.B.; Guedes, I.A.; Karl, A.L.M.; Dardenne, L.E. Highly Flexible Ligand Docking: Benchmarking of the DockThor Program on the LEADS-PEP Protein–Peptide Data Set. J. Chem. Inf. Model. 2020, 60, 667–683. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein–Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, 1–5. [Google Scholar] [CrossRef]
- Fang, N.; Casida, J.E. Cubé Resin Insecticide: Identification and Biological Activity of 29 Rotenoid Constituents. J. Agric. Food Chem. 1999, 47, 2130–2136. [Google Scholar] [CrossRef]
- Rowlands, J.C.; Casida, J.E. NADH: Ubiquinone Oxidoreductase Inhibitors Block Induction of Ornithine Decarboxylase Activity in MCF-7 Human Breast Cancer Cells. Pharmacol. Toxicol. 1998, 83, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Darrouzet, E.; Issartel, J.; Dupuis, A. The 49-KDa Subunit of NADH-Ubiquinone Oxidoreductase (Complex I) Is Involved in the Binding of Piericidin and Rotenone, Two Quinone-Related Inhibitors. FEBS Lett. 1998, 431, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Zong, S.; Wu, M.; Gu, J.; Yang, M. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell 2017, 170, 1247–1257.e12. [Google Scholar] [CrossRef] [Green Version]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [Green Version]
- Wallace, I.; O’Sullivan, O.; Higgins, D.; Notredame, C. M-Coffee: Combining Multiple Sequence Alignment Methods with T-Coffee. Nucleic Acids Res. 2006, 34, 1692–1699. [Google Scholar] [CrossRef] [PubMed]
- Real, F.; Vidal, R.O.; Carazzolle, M.F.; Mondego, J.M.C.; Costa, G.G.L.; Herai, R.H.; Würtele, M.; de Carvalho, L.M.; e Ferreira, R.C.; Mortara, R.A.; et al. The Genome Sequence of Leishmania (Leishmania) Amazonensis: Functional Annotation and Extended Analysis of Gene Models. DNA Res. 2013, 20, 567–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridges, H.R.; Fedor, J.G.; Blaza, J.N.; Di Luca, A.; Jussupow, A.; Jarman, O.D.; Wright, J.J.; Agip, A.-N.A.; Gamiz-Hernandez, A.P.; Roessler, M.M.; et al. Structure of Inhibitor-Bound Mammalian Complex I. Nat. Commun. 2020, 11, 5261. [Google Scholar] [CrossRef]
- Dufe, V.T.; Ingner, D.; Heby, O.; Khomutov, A.R.; Persson, L.; Al-karadaghi, S.; We, M.; African, W.; Key, C.; African, W.; et al. A Structural Insight into the Inhibition of Human and Leishmania Donovani Ornithine Decarboxylases by 1-Amino-Oxy-3-Aminopropane. Biochem. J. 2007, 405, 261–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, C.; Wang, O.; Strutz, S.E.; González-Salazar, C.; Sánchez-Cordero, V.; Sarkar, S. Climate Change and Risk of Leishmaniasis in North America: Predictions from Ecological Niche Models of Vector and Reservoir Species. PLoS Negl. Trop. Dis. 2010, 4, e585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerhã, C.; Lee, S.K.; Kosmeder, J.W.; Moriarty, R.M.; Hamel, E.; Mehta, R.G.; Moon, R.C.; Pezzuto, J.M. Regulation of Ornithine Decarboxylase Induction by Deguelin, a Natural Product Cancer Chemopreventive Agent1. Cancer Res. 1997, 57, 3429–3436. [Google Scholar]
- Gerhäuser, C.; Mar, W.; Lee, S.K.; Suh, N.; Luo, Y.; Kosmeder, J.; Luyengi, L.; Harry, H.S.; Fong, A.; KingHorn, D.; et al. Mehta, Andreas Constantinou, R.C.M.& J.M.P. Rotenoids Mediate Potent Cancer Chemopreventive Activity through Transcriptional Regulation of Ornithine Decarboxylase. Nat. Med. 1995, 3, 260–266. [Google Scholar]
- Robledo, S.M.; Echeverri, F. Preparation of Rotenone Derivatives and in Vitro Analysis of Their Antimalarial, Antileishmanial and Selective Cytotoxic Activities. Molecules 2014, 19, 18911–18922. [Google Scholar] [CrossRef] [Green Version]
- Heby, O.; Roberts, S.C.; Ullman, B. Polyamine Biosynthetic Enzymes as Drug Targets in Parasitic Protozoa. Biochem. Soc. Trans. 2003, 31, 415–419. [Google Scholar] [CrossRef] [Green Version]
- Heby, O.; Persson, L.; Rentala, M. Targeting the Polyamine Biosynthetic Enzymes: A Promising Approach to Therapy of African Sleeping Sickness, Chagas ’ Disease, and Leishmaniasis Review Article. Aminoacids 2007, 33, 359–366. [Google Scholar] [CrossRef]
- Scotti, L.; Ishiki, H.; Mendonca, F.J.B.; Silva, M.S.; Scotti, M.T. In-Silico Analyses of Natural Products on Leishmania Enzyme Targets. Mini-Reviews Med. Chem. 2015, 15, 253–269. [Google Scholar] [CrossRef]
- Singh, S.; Mukherjee, A.; Khomutov, A.R.; Persson, L.; Heby, O.; Chatterjee, M.; Madhubala, R. Antileishmanial Effect of 3-Aminooxy-1-Aminopropane Is Due to Polyamine Depletion. Antimicrob. Agents Chemother. 2007, 51, 528–534. [Google Scholar] [CrossRef] [Green Version]
- Boitz, J.M.; Yates, P.A.; Kline, C.; Gaur, U.; Wilson, M.E.; Ullman, B.; Roberts, S.C. Leishmania Donovani Ornithine Decarboxylase Is Indispensable for Parasite Survival in the Mammalian Host. Infect. Immun. 2009, 77, 756–763. [Google Scholar] [CrossRef] [Green Version]
- Gerner, E.W.; Jr, F.L.M.; Family, C. Polyamines And Cancer: Old Molecules, New Understanding. Nat. Rev. Cancer 2004, 4, 781–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilari, A.; Fiorillo, A.; Baiocco, P.; Poser, E.; Angiulli, G.; Colotti, G. Targeting Polyamine Metabolism for Finding New Drugs Against Leishmaniasis: A Review. Mini-Reviews Med. Chem. 2015, 15, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Vannier-santos, M.A.; Menezes, D.; Oliveira, M.F.; De Mello, F.G. The Putrescine Analogue 1, 4-Diamino-2-Butanone Affects Polyamine Synthesis, Transport, Ultrastructure and Intracellular Survival in Leishmania Amazonensis. Microbiology 2008, 154, 3104–3111. [Google Scholar] [CrossRef] [Green Version]
- Kern, A.D.; Oliveira, M.A.; Coffino, P.; Hackert, M.L. Structure of Mammalian Ornithine Decarboxylase at 1. 6 Å Resolution: Stereochemical Implications of PLP-Dependent Amino Acid Decarboxylases. Structure 1999, 7, 567–581. [Google Scholar] [CrossRef] [Green Version]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved Protein Structure Prediction Using Potentials from Deep Learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]








| Drugs | Mechanism of Action |
|---|---|
| Antimonials | Antimonial complexes are administrated as Antimony (V) form which is reduced to Sb (III) in the lysosome. The reduced form inhibits some of these three targets; trypanothione, trypanothione reductase, or nucleoside topoisomerase [15]. |
| Amphotericin B (liposomal) | Amphotericin B binds to ergosterol (sterol that is the main component of fungal and some protists cell membranes, performing the same function as cholesterol in animal cells) present in the cell membrane of parasites of the genus Leishmania. This binding destabilizes the membrane causing the release of that intracellular ions triggering the cell death. When administered through liposomes, the stability and specificity of amphotericin B is improved [16,17]. |
| Miltefosine | Miltefosine is a phosphocholine analogue that inhibits the synthesis of phosphatidylcholine of the parasite, as well as the cytochrome c oxidase affecting the mitochondrial membrane potential [18]. |
| Paromomycin | Paromomycin is an aminoglycoside antibiotic that binds to the 30S subunit of the ribosome, thus that inhibits protein synthesis [19]. Also, it has been proposed that could alter mitochondrial membrane potential and inhibition of mitochondrial respiration chain [20]. |
| Pentamidine | Pentamidine interferes with polyamine synthesis, RNA polymerase activity, enters the protozoal cell binding to transfer RNA, and prevents the synthesis of protein, nucleic acids, phospholipids, and folate. Additionally, it is known to be an anti-inflammatory agent, xenobiotic, and an antagonist of the NMDA receptor, histone acetyltransferase, and calmodulin [21]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vicente-Barrueco, A.; Román, Á.C.; Ruiz-Téllez, T.; Centeno, F. In Silico Research of New Therapeutics Rotenoids Derivatives against Leishmania amazonensis Infection. Biology 2022, 11, 133. https://doi.org/10.3390/biology11010133
Vicente-Barrueco A, Román ÁC, Ruiz-Téllez T, Centeno F. In Silico Research of New Therapeutics Rotenoids Derivatives against Leishmania amazonensis Infection. Biology. 2022; 11(1):133. https://doi.org/10.3390/biology11010133
Chicago/Turabian StyleVicente-Barrueco, Adrián, Ángel Carlos Román, Trinidad Ruiz-Téllez, and Francisco Centeno. 2022. "In Silico Research of New Therapeutics Rotenoids Derivatives against Leishmania amazonensis Infection" Biology 11, no. 1: 133. https://doi.org/10.3390/biology11010133