The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Metallophores Produced by Staphylococcus aureus
2.1. Siderophores
2.1.1. Ferric Uptake Regulator (Fur)
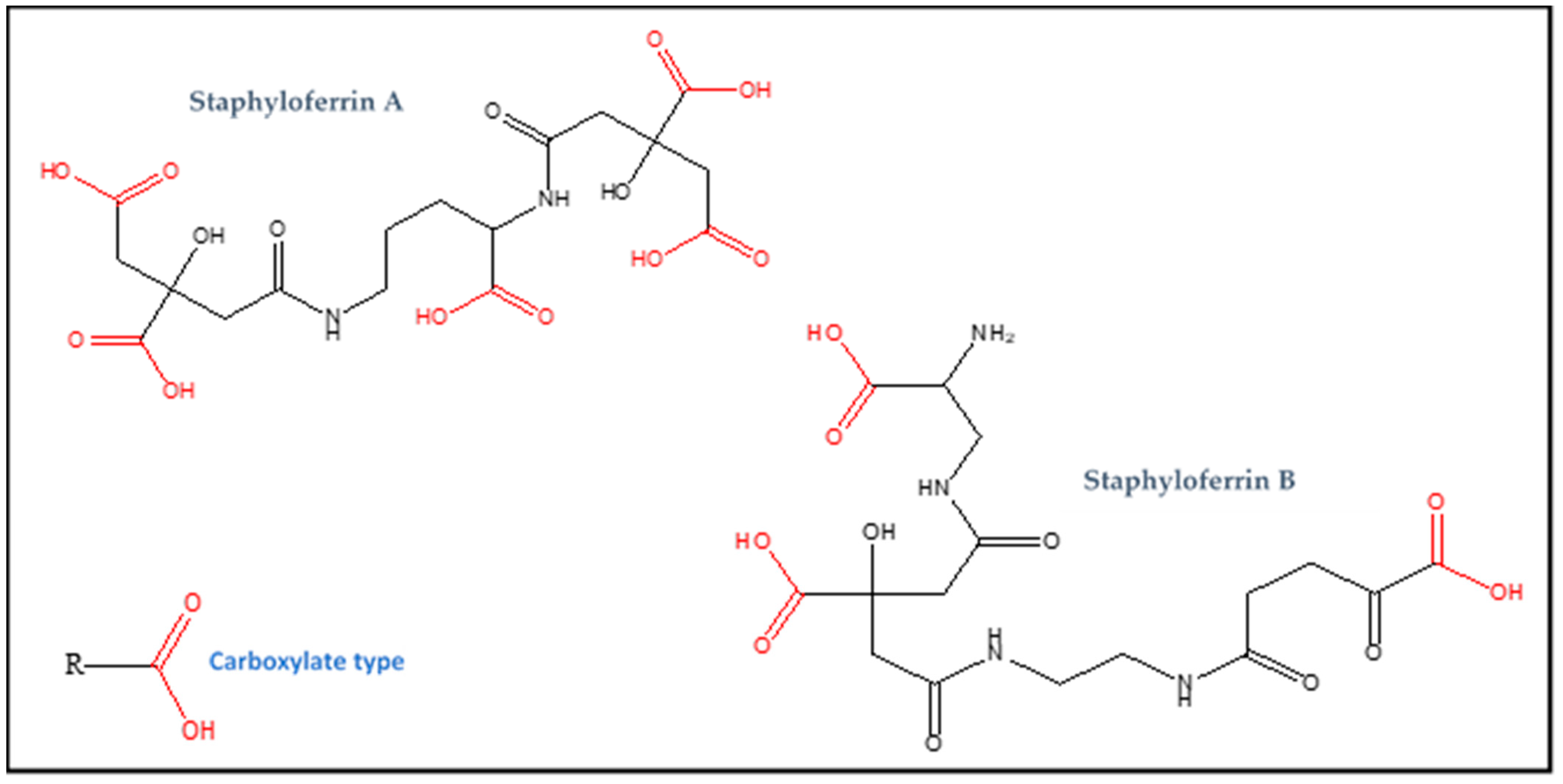
2.1.2. Staphyloferrin A
2.1.3. Staphyloferrin B
2.1.4. Staphylobactin
2.1.5. Aureochelin
2.1.6. Xenosiderophores Uptake by S. aureus
2.1.7. The Virulence of Staphylococcal Siderophores
2.2. Additional Metal Acquiring Systems
2.2.1. Metallophore Staphylopine
2.2.2. Staphylopine Synthesis
2.2.3. Staphylopine as Zincophore
2.2.4. Important Features of Cnt-Staphylopine System
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coates, R.; Moran, J.; Horsburgh, M.J. Staphylococci: Colonizers and pathogens of human skin. Future Microbiol. 2014, 9, 75–91. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Van Hal, S.J.; Jensen, S.O.; Vaska, V.L.; Espedido, B.A.; Paterson, D.L.; Gosbell, I.B. Predictors of mortality in Staphylococcus aureus Bacteremia. Clin. Microbiol. Rev. 2012, 25, 362–386. [Google Scholar] [CrossRef] [Green Version]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 322, 207–228. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Shockman, G.D.; Barren, J.F. Structure, Function, and Assembly of Cell Walls of Gram-Positive Bacteria. Annu. Rev. Microbiol. 1983, 37, 501–527. [Google Scholar] [CrossRef]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus Toxins and Their Molecular Activity in Infectious Diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, M.E.; McLoughlin, R.M. Host–bacterial crosstalk determines Staphylococcus aureus nasal colonization. Trends Microbiol. 2016, 24, 872–886. [Google Scholar] [CrossRef]
- Torres Salazar, B.O.; Heilbronner, S.; Peschel, A.; Krismer, B. Secondary Metabolites Governing Microbiome Interaction of Staphylococcal Pathogens and Commensals. Microb. Physiol. 2021, 31, 198–216. [Google Scholar] [CrossRef]
- Garcia, Y.M.; Barwinska-Sendra, A.; Tarrant, E.; Skaar, E.P.; Waldron, K.J.; Kehl-Fie, T.E. A Superoxide Dismutase Capable of Functioning with Iron 472 or Manganese Promotes the Resistance of Staphylococcus aureus to Calprotectin and 473 Nutritional Immunity. PLoS Pathog. 2017, 13, e1006125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, N.D.; Skaar, E.P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerinot, M.L. Microbial iron transport. Annu. Rev. Microbiol. 1994, 48, 743–772. [Google Scholar] [CrossRef] [PubMed]
- Courcol, R.J.; Trivier, D.; Bissinger, M.C.; Martin, G.R.; Brown, M.R. Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infect. Immun. 1997, 65, 1944–1948. [Google Scholar] [CrossRef] [Green Version]
- Dale, S.E.; Sebulsky, M.T.; Heinrichs, D.E. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 2004, 186, 8356–8362. [Google Scholar] [CrossRef] [Green Version]
- Grim, K.P.; San Francisco, B.; Radin, J.N.; Brazel, E.B.; Kelliher, J.L.; Párraga Solórzano, P.K.; Kim, P.C.; McDevitt, C.A.; Kehl-Fie, T.E. The Metallophore Staphylopine Enables Staphylococcus aureus To Compete with the Host for Zinc and Overcome Nutritional Immunity. mBio 2017, 8, e01281-17. [Google Scholar] [CrossRef] [Green Version]
- Remy, L.; Carrière, M.; Derré-Bobillot, A.; Martini, C.; Sanguinetti, M.; Borezée-Durant, E. The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Mol. Microbiol. 2013, 87, 730–743. [Google Scholar] [CrossRef]
- Hiron, A.; Posteraro, B.; Carrière, M.; Remy, L.; Delporte, C.; La Sorda, M.; Sanguinetti, M.; Juillard, V.; Borezée-Durant, E. A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection. Mol. Microbiol. 2010, 77, 1246–1260. [Google Scholar] [CrossRef]
- Skaar, E.P.; Schneewind, O. Iron-regulated surface determinants (Isd) of 520 Staphylococcus aureus: Stealing iron from heme. Microbes Infect. 2004, 6, 390–397. [Google Scholar] [CrossRef]
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus Perception in Bacterial Signal-Transducing 529 Histidine Kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gkouvatsos, K.; Papanikolaou, G.; Pantopoulos, K. Regulation of iron transport and the role of transferrin. Biochim. Biophys. Acta 2012, 1820, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, W.; Wu, S.; Gao, H. Recent Advances in the Siderophore Biology of Shewanella. Front. Microbiol. 2022, 13, 823758. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995, 82, 969–974. [Google Scholar] [CrossRef]
- Emerson, D.; Roden, E.; Twining, B.S. The microbial ferrous wheel: Iron cycling in terrestrial, freshwater, and marine environments. Front. Microbiol. 2012, 3, 383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, J.; Özkaya, O.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef]
- Sebulsky, M.T.; Hohnstein, D.; Hunter, M.D.; Heinrichs, D.E. Identification and characterization of a membrane permease ivolved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 2000, 182, 4394–4400. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Cassat, J.E.; Skaar, E.P. Iron in infection and immunity. Cell Host Microbe 2013, 13, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Challis, G.L. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chem. Bio. Chem. 2005, 6, 601–661. [Google Scholar] [CrossRef]
- Carroll, C.S.; Moore, M.M. Ironing out siderophore biosynthesis: A review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 356–381. [Google Scholar] [CrossRef] [PubMed]
- Crosa, J.H.; Walsh, C.T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 223–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.Y.; Janes, B.K.; Passalacqua, K.D.; Pfleger, B.F.; Bergman, N.H.; Liu, H.; Håkansson, K.; Somu, R.V.; Aldrich, C.C.; Cendrowski, S.; et al. Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J. Bacteriol. 2007, 189, 1698–1710. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.C.; Robinson, A.K.; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, J.R.; Heinrichs, D.E. Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol. Rev. 2015, 39, 592–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faraldo-Gómez, J.D.; Sansom, M.S.P. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 2003, 4, 105–116. [Google Scholar] [CrossRef]
- Schalk, I.J.; Guillon, L. Fate of ferrisiderophores after import across bacterial outer membranes: Different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 2013, 44, 1267–1277. [Google Scholar] [CrossRef]
- Ganne, G.; Brillet, K.; Basta, B.; Roche, B.; Hoegy, F.; Gasser, V.; Schalk, I.J. Iron release from the siderophore pyoverdine in Pseudomonas aeruginosa involves three new actors: FpvC, FpvG, and FpvH. ACS Chem. Biol. 2017, 12, 1056–1065. [Google Scholar] [CrossRef]
- Krewulak, K.D.; Vogel, H.J. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta 2008, 1778, 1781–1804. [Google Scholar] [CrossRef] [Green Version]
- Imperi, F.; Tiburzi, F.; Visca, P. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 20440–20445. [Google Scholar] [CrossRef]
- Lin, H.; Fischbach, M.A.; Liu, D.R.; Walsh, C.T. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 2005, 127, 11075–11084. [Google Scholar] [CrossRef] [Green Version]
- Torres, V.J.; Pishchany, G.; Humayun, M.; Schneewind, O.; Skaar, E.P. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 2006, 188, 8421–8429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baichoo, N.; Wang, T.; Ye, R.; Helmann, J.D. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 2002, 45, 1613–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pishchany, G.; Dickey, S.E.; Skaar, E.P. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect. Immun. 2009, 77, 2624–2634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reniere, M.L.; Skaar, E.P. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 2008, 69, 1304–1315. [Google Scholar] [CrossRef] [Green Version]
- Friedman, D.B.; Stauff, D.L.; Pishchany, G.; Whitwell, C.W.; Torres, V.J.; Skaar, E.P. Staphylococcus aureus Redirects Central Metabolism to Increase Iron Availability. PLoS Pathog. 2006, 2, e87. [Google Scholar] [CrossRef]
- Chavakis, T.; Hussain, M.; Kanse, S.M.; Peters, G.; Bretzel, R.G.; Flock, J.-I.; Herrmann, M.; Preissner, K.T. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 2002, 8, 687–693. [Google Scholar] [CrossRef]
- Johnson, M.; Cockayne, A.; Morrissey, J.A. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immun. 2008, 76, 1756–1765. [Google Scholar] [CrossRef] [Green Version]
- Bagg, A.; Neilands, J.B. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator in an iron transport operon in Escherichia coli. Biochemistry 1987, 26, 5471–5477. [Google Scholar] [CrossRef]
- Horsburgh, M.J.; Clements, M.O.; Crossley, H.; Ingram, E.; Foster, S.J. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 2001, 69, 744–3754. [Google Scholar] [CrossRef]
- Conroy, B.S.; Grigg, J.C.; Kolesnikov, M.; Morales, L.D.; Murphy, M.E. Staphylococcus aureus heme and siderophore-iron acquisition pathways. Biometals 2019, 32, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Beasley, F.C.; Vinés, E.D.; Grigg, J.C.; Zheng, Q.; Liu, S.; Lajoie, G.A.; Murphy, M.E.P.; Heinrichs, D.E. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol. Microbiol. 2009, 72, 947–963. [Google Scholar] [CrossRef] [PubMed]
- Beasley, F.C.; Heinrichs, D.E. Siderophore-mediated iron acquisition in the staphylococci. J. Inorg. Biochem. 2010, 104, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Hannauer, M.; Sheldon, J.R.; Heinrichs, D.E. Involvement of major facilitator superfamily proteins SfaA and SbnD in staphyloferrin secretion in Staphylococcus aureus. FEBS Lett. 2015, 589, 730–737. [Google Scholar] [CrossRef] [Green Version]
- Cotton, J.L.; Tao, J.; Balibar, C.J. Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. Biochemistry 2009, 48, 1025–1035. [Google Scholar] [CrossRef]
- Sheldon, J.R.; Marolda, C.L.; Heinrichs, D.E. TCA cycle activity in Staphylococcus aureus is essential for ironregulated synthesis of staphyloferrin A, but not staphyloferrin B: The benefit of a second citrate synthase. Mol. Microbiol. 2014, 92, 824–839. [Google Scholar] [CrossRef]
- Hannauer, M.; Arifin, A.J.; Heinrichs, D.E. Involvement of reductases IruO and NtrA in iron acquisition by Staphylococcus aureus. Mol. Microbiol. 2015, 96, 1192–1210. [Google Scholar] [CrossRef]
- Perera, V.R.; Newton, G.L.; Pogliano, K. Bacillithiol: A key protective thiol in Staphylococcus aureus. Expert Rev. Anti-infect. Ther. 2015, 13, 1089–1107. [Google Scholar] [CrossRef] [Green Version]
- Dale, S.E.; Doherty-Kirby, A.; Lajoie, G.; Heinrichs, D.E. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: Identification and characterization of genes involved in production of a siderophore. Infect. Immun. 2004, 72, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Cheung, J.; Beasley, F.C.; Liu, S.; Lajoie, G.A.; Heinrichs, D.E. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 2009, 74, 594–608. [Google Scholar] [CrossRef]
- Allard, M.; Moisan, H.; Brouillette, E.; Gervais, A.L.; Jacques, M.; Lacasse, P.; Diarra, M.S.; Malouin, F. Transcriptional modulation of some Staphyloccocus aureus iron-regulated genes during growth in vitro and in a tissue cage model in vivo. Microbes Infect. 2006, 8, 1679–1690. [Google Scholar] [CrossRef] [PubMed]
- Kobylarz, M.J.; Grigg, J.C.; Liu, Y.; Lee, M.S.F.; Heinrichs, D.E.; Murphy, M.E.P. Deciphering the substrate specificity of SbnA, the enzyme catalyzing the first step in Staphyloferrin B biosynthesis. Biochemistry 2016, 55, 927–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laakso, H.A.; Marolda, C.L.; Pinter, T.B.; Stillman, M.J.; Heinrichs, D.E. A heme-responsive regulator controls synthesis of staphyloferrin B in Staphylococcus aureus. J. Biol. Chem. 2016, 291, 29–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstraete, M.M.; Perez-Borrajero, C.; Brown, K.L.; Heinrichs, D.E.; Murphy, M.E.P. SbnI is a free serine kinase that generates O-phospho-L-serine for staphyloferrin B biosynthesis in Staphylococcus aureus. J. Biol. Chem. 2018, 293, 6147–6160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speziali, C.D.; Dale, S.E.; Henderson, J.A.; Vine’s, E.D.; Heinrichs, D.E. Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J. Bacteriol. 2006, 188, 2048–2055. [Google Scholar] [CrossRef] [Green Version]
- Grigg, J.C.; Cheung, J.; Heinrichs, D.E.; Murphy, M.E.P. Specificity of staphyloferrin B recognition by the SirA receptor from Staphylococcus aureus. J. Biol. Chem. 2010, 285, 34579–34588. [Google Scholar] [CrossRef] [Green Version]
- Wysocki, P.; Lisiecki, P.; Mikucki, J. Receptors for endogenous and heterogeneous siderophores in Staphylococcus aureus B 471. Pol. J. Microbiol. 2005, 54, 97–103. [Google Scholar]
- Sebulsky, M.T.; Speziali, C.D.; Shilton, B.H.; Edgell, D.R.; Heinrichs, D.E. FhuD1, a ferric hydroxamate-binding lipoprotein in Staphylococcus aureus: A case of gene duplication and lateral transfer. J. Biol. Chem. 2004, 279, 53152–53159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera, G.; Xiong, A.; Uebel, M.; Singh, V.K.; Jayaswal, R.K. Molecular characterization of the iron-hydroxamate uptake system in Staphylococcus aureus. Appl. Environ. Microbiol. 2001, 67, 1001–1003. [Google Scholar] [CrossRef] [Green Version]
- Sebulsky, M.T.; Heinrichs, D.E. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 2001, 183, 4994–5000. [Google Scholar] [CrossRef] [Green Version]
- Sebulsky, M.T.; Shillton, B.H.; Speziali, D.; Heinrichs, D.E. The role of FhuD2 in iron(III)-hydroxamate transport in Staphylococcus aureus. Demonstration that FhuD2 binds iron(III)-hydroxamamtes but with minimal conformational change and implications of mutations on transport. J. Biol. Chem. 2003, 278, 49890–49900. [Google Scholar] [CrossRef] [PubMed]
- Beasley, F.C.; Marolda, C.L.; Cheung, J.; Buac, S.; Heinrichs, D.E. Staphylococcus aureus transporters Hts, Sir, and Sst capture iron liberated from human transferrin by staphyloferrin A, staphyloferrin B, and catecholamine stress hormones, respectively, and contribute to virulence. Infect. Immun. 2011, 79, 2345–2355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrissey, J.A.; Cockayne, A.; Hill, P.J.; Williams, P. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 2000, 68, 6281–6288. [Google Scholar] [CrossRef] [PubMed]
- Beasley, F.C.; Cheung, J.; Heinrichs, D.E. Mutation of L-2,3- diaminopropionic acid synthase genes blocks staphyloferrin B synthesis in Staphylococcus aureus. BMC Microbiol. 2011, 11, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozalska, B.; Lisiecki, P.; Sadowska, B.; Mikucki, J.; Rudnicka, W. The virulence of Staphylococcus aureus isolates differing by siderophore production. Acta Microbiol. Pol. 1998, 47, 185–194. [Google Scholar] [PubMed]
- Lebrette, H.; Brochier-Armanet, C.; Zambelli, B.; de Reuse, H.; Borezée-Durant, E.; Ciurli, S.; Cavazza, C. Promiscuous nickel import in human pathogens: Structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins. Structure 2014, 22, 1421–1432. [Google Scholar] [CrossRef] [Green Version]
- Eitinger, T.; Suhr, J.; Moore, L.; Smith, J.A. Secondary transporters for nickel and cobalt ions: Theme and variations. Biometals 2005, 18, 399–405. [Google Scholar] [CrossRef]
- Lebrette, H.; Borezée-Durant, E.; Martin, L.; Richaud, P.; Erba, E.B.; Cavazza, C. Novel insights into nickel import in Staphylococcus aureus: The positive role of free histidine and structural characterization of a new thiazolidine- type nickel chelator. Metallomics 2015, 7, 613–621. [Google Scholar] [CrossRef]
- Ghssein, G.; Brutesco, C.; Ouerdane, L.; Fojcik, C.; Izaute, A.; Wang, S.; Hajjar, C.; Lobinski, R.; Lemaire, D.; Richaud, P.; et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 2016, 352, 1105–1109. [Google Scholar] [CrossRef]
- Ghssein, G.; Matar, S.F. Chelating Mechanisms of Transition Metals by Bacterial Metallophores “Pseudopaline and Staphylopine”: A Quantum Chemical Assessment. Computation 2018, 6, 56. [Google Scholar] [CrossRef] [Green Version]
- Hiron, A.; Borezee-Durant, E.; Piard, J.C.; Juillard, V. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 2007, 189, 5119–5129. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fu, Y.; Lee, J.C.; Hooper, D.C. Staphylococcus aureus NorD, a putative efflux pump coregulated with the Opp1 oligopeptide permease, contributes selectively to fitness in vivo. J. Bacteriol. 2012, 194, 6586–6593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Zhang, Y.; Chen, W.; Gu, T.; Zhang, S.Y.; Ji, Q. Mechanistic insights into staphylopine-mediated metal acquisition. Proc. Natl. Acad. Sci. USA 2018, 115, 3942–3947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, J.A.; Foster, S.J. zur: A Zn(21)-responsive regulatory element of Staphylococcus aureus. Microbiology 2001, 147, 1259–1266. [Google Scholar] [CrossRef] [Green Version]
- Fojcik, C.; Arnoux, P.; Ouerdane, L.; Aigle, M.; Alfonsi, L.; Borezée-Durant, E. Independent and Cooperative Regulation of Staphylopine Biosynthesis and Trafficking by Fur and Zur. Mol. Microbiol. 2018, 108, 159–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajjar, C.; Fanelli, R.; Laffont, C.; Brutesco, C.; Cullia, G.; Tribout, M.; Nurizzo, D.; Borezée-Durant, E.; Voulhoux, R.; Pignol, D.; et al. Control by Metals of Staphylopine Dehydrogenase Activity during Metallophore Biosynthesis. Am. Chem. Soc. 2019, 141, 5555–5562. [Google Scholar] [CrossRef]
- Corbin, B.D.; Seeley, E.H.; Raab, A.; Feldmann, J.; Miller, M.R.; Torres, V.J.; Anderson, K.L.; Dattilo, B.M.; Dunman, P.M.; Gerads, R.; et al. Metal chelation and inhibition of bacterial growth tissue abscesses. Science 2008, 319, 962–965. [Google Scholar] [CrossRef]
- Kehl-Fie, T.E.; Zhang, Y.; Moore, J.L.; Farrand, A.J.; Hood, M.I.; Rathi, S.; Chazin, W.J.; Caprioli, R.M.; Skaar, E.P. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect. Immun. 2013, 81, 3395–3405. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Hooper, D.C. Intracellular accumulation of staphylopine impairs the fitness of Staphylococcus aureus cntE mutant. FEBS Lett. 2019, 593, 1213–1222. [Google Scholar] [CrossRef]
- Grim, K.P.; Radin, J.N.; Solórzano, P.K.P.; Morey, J.R.; Frye, K.A.; Ganio, K.; Neville, S.L.; McDevitt, C.A.; Kehl-Fie, T.E. Intracellular Accumulation of Staphylopine Can Sensitize Staphylococcus aureus to Host-Imposed Zinc Starvation by Chelation-Independent Toxicity. J. Bacteriol. 2020, 202, 00014–00020. [Google Scholar] [CrossRef]
- Luo, S.; Ju, Y.; Zhou, J.; Gu, Q.; Xu, J.; Zhou, H. Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus. Int. J. Biol. Macromol. 2019, 135, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Luo, S.; Ju, Y.; Ding, P.; Xu, J.; Gu, Q.; Zhou, H. Structural insights into the ligand recognition and catalysis of the key aminobutanoyltransferase CntL in staphylopine biosynthesis. FASEB J. 2021, 5, 21575. [Google Scholar] [CrossRef] [PubMed]
- Abideen, Z.U.; Ahmad, A.; Usman, M.; Majaz, S.; Ali, W.; Noreen, S.; Mahmood, T.; Nouroz, F. Dynamics and conformational propensities of staphylococcal CntA. J. Biomol. Struct. Dyn. 2021, 39, 4923–4935. [Google Scholar] [CrossRef] [PubMed]
- Vinué, L.; Hooper, D.C. Rsp activates expression of the Cnt system in Staphylococcus aureus. BMC Microbiol. 2020, 20, 327. [Google Scholar] [CrossRef]
- Kotecka, K.; Kawalek, A.; Kobylecki, K.; Bartosik, A.A. The AraC-Type Transcriptional Regulator GliR (PA3027) Activates Genes of Glycerolipid Metabolism in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 5066. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Wang, Z.; Wu, Z.; Wang, Y.; Tang, N.; Xu, X.; Zhao, S.; Chen, W.; Ji, Q. Programmable adenine deamination in bacteria using a Cas9-adenine-deaminase fusion. Chem. Sci. 2020, 6, 1657–1664. [Google Scholar] [CrossRef]

| Name | Type (Functional Group) | Molecular Formula | Molecular Weight (Da) | Specific for |
|---|---|---|---|---|
| Staphyloferrin A | Carboxylate (RCOOH) | C17H24N2O14 | 480 | Iron |
| Staphyloferrin B | Carboxylate (RCOOH) | C16H24N4O11 | 448 | Iron |
| Staphylobactin | Hydroxamate (R-CO-NH-OH) | unknown | 822 | Iron |
| Aureochelin | hydroxamate and catechol (R-CO-NH-OH and C6H4(OH)2) | unknown | 577 | Iron |
| Staphylopine | opine (amine and carboxylic acid) | C13H19N4O6− | 327 | broad-spectrum metallophore (nickel, zinc, cobalt, iron and copper) |
| Name | Operon | Metal Uptake Regulator | Membrane Transporter |
|---|---|---|---|
| Staphyloferrin A | sfaABCD | Fur | HtsABC |
| Staphyloferrin B | sbn (sbnABCDEFGHI) | Fur | SirABC |
| Staphylobactin | sbn | Fur | SirABC |
| Staphylopine | cnt (cntKLMABCDFE) | Fur and Zur | CntB and CntC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghssein, G.; Ezzeddine, Z. The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review. Biology 2022, 11, 1525. https://doi.org/10.3390/biology11101525
Ghssein G, Ezzeddine Z. The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review. Biology. 2022; 11(10):1525. https://doi.org/10.3390/biology11101525
Chicago/Turabian StyleGhssein, Ghassan, and Zeinab Ezzeddine. 2022. "The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review" Biology 11, no. 10: 1525. https://doi.org/10.3390/biology11101525






