Transcriptomic Analysis of Hepatitis B Infected Liver for Prediction of Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohorts and Microarray Data Processing
2.2. Evaluation of Protein-Level Expression
2.3. Bioinformatics Analyses
2.4. Statistical Analyses
3. Results
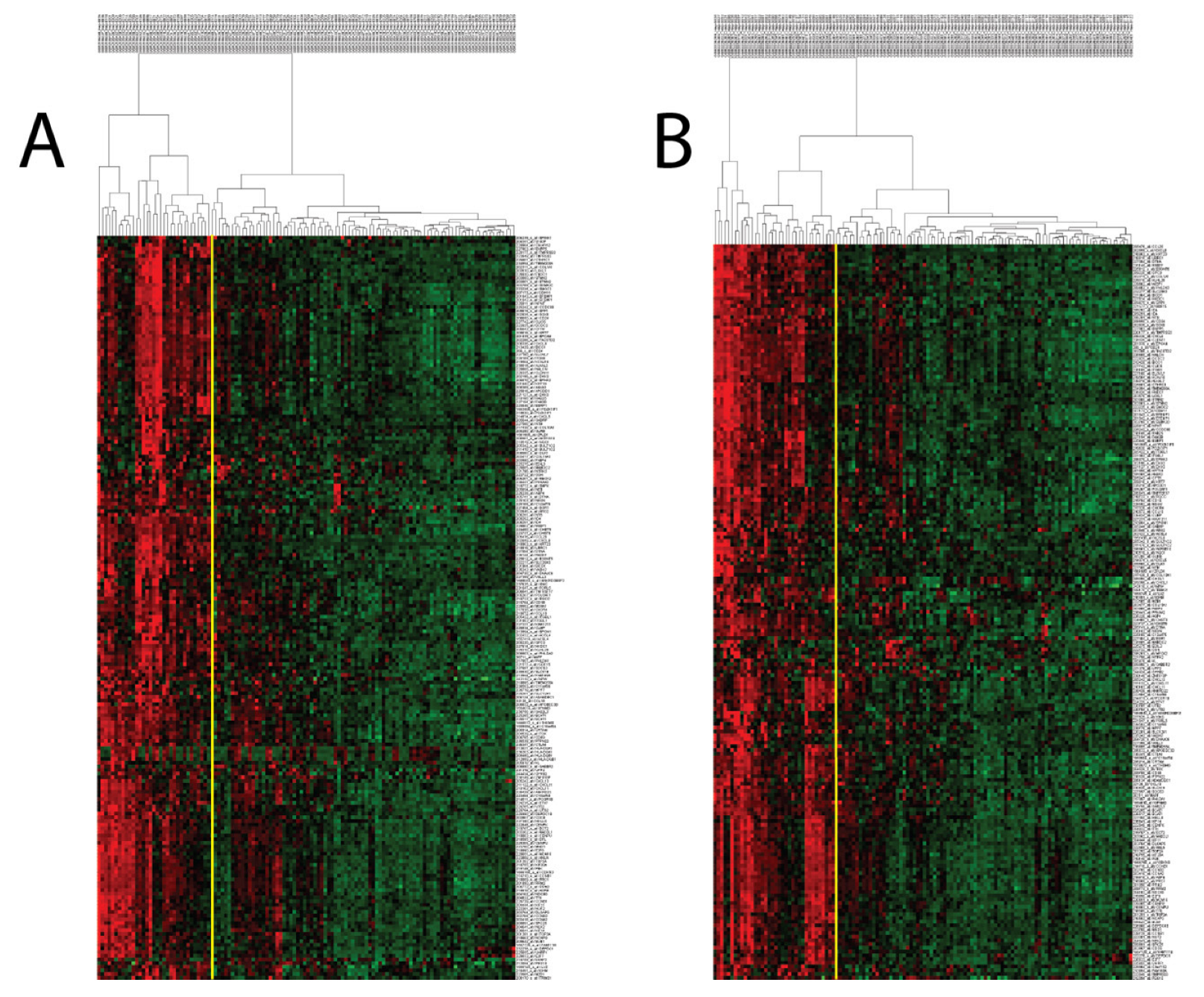
3.1. Identifying Transcriptomic Sub-Groups among CHB Patient Liver Samples
3.2. PCM1 Signature Identifies a “Proliferation, Cancer and M1 Macrophage” Related Phenotype
3.3. Transcriptomic Changes Suggest a Higher Risk of Malignant Transformation in PCM1-U Group
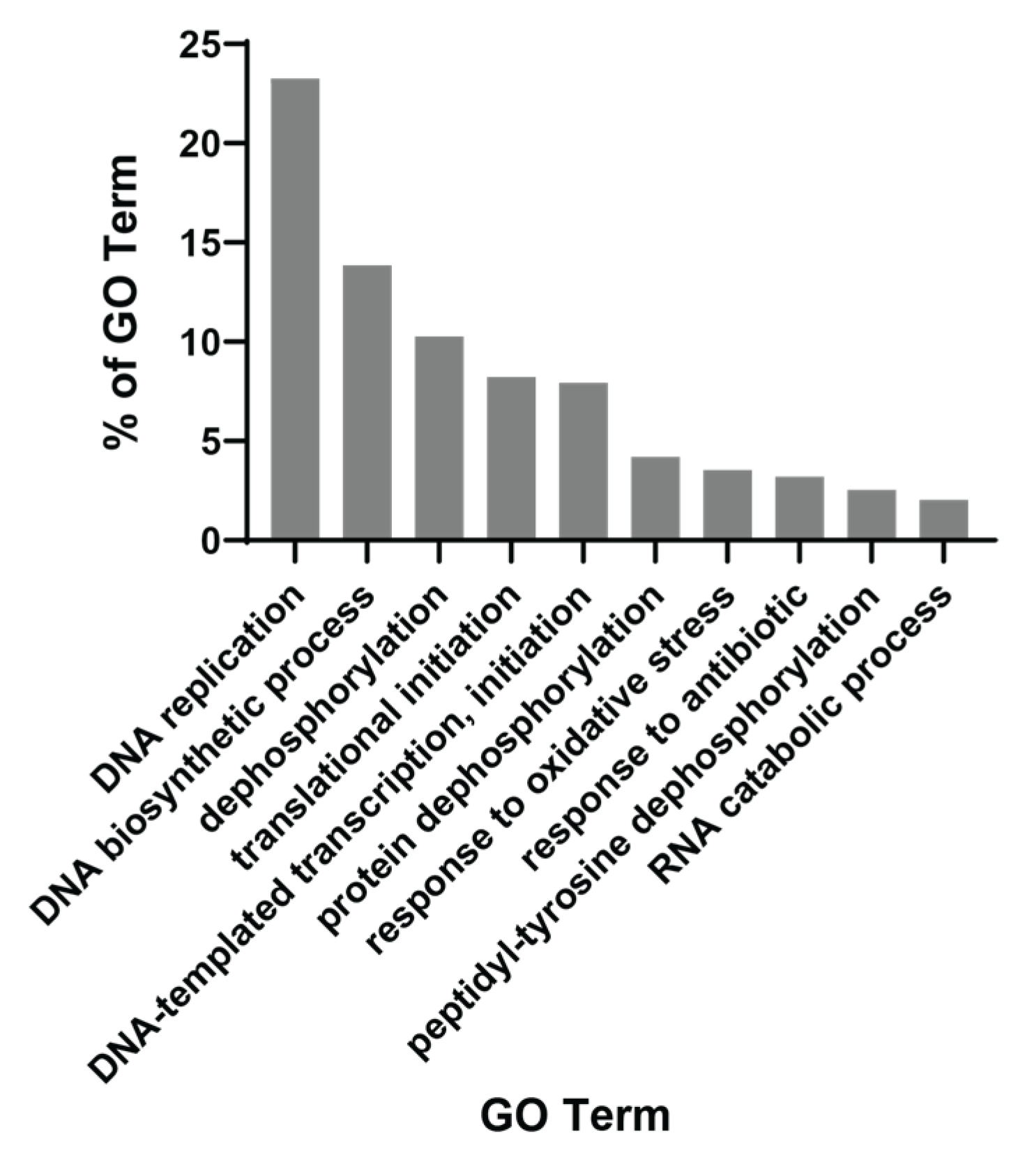
3.4. The Majority of PCM1 Genes Are Related to Mitosis and Immune Response
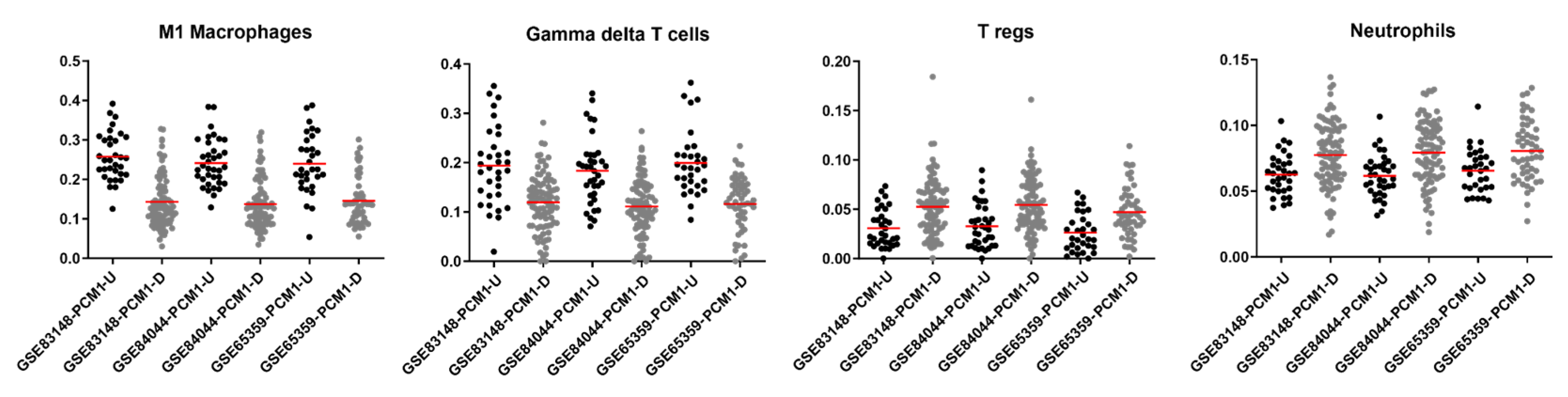
3.5. Specific Immune Cell Types Are Prominent in PCM1 Groups
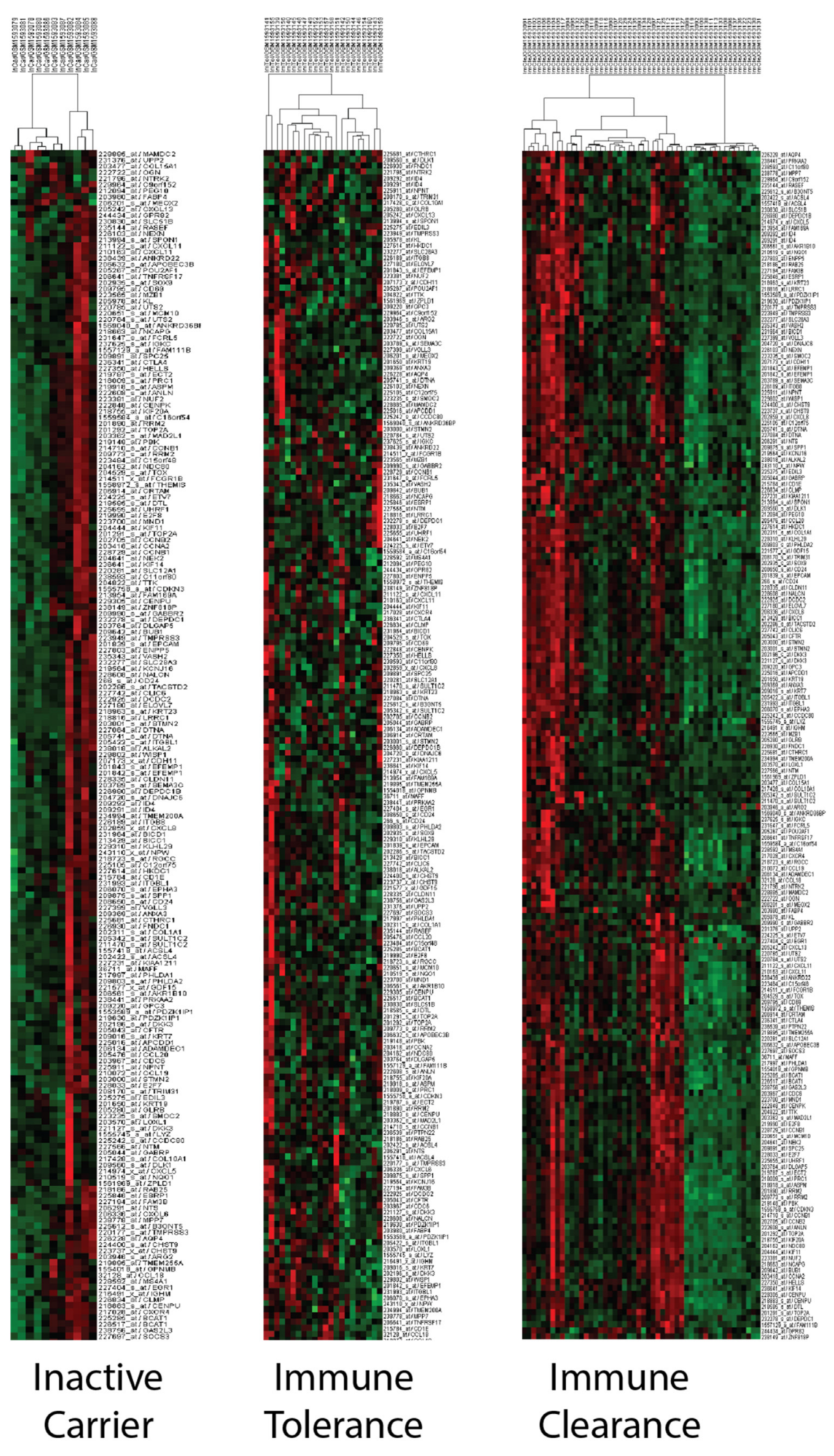
3.6. PCM1 Groups Show Differences in ALT, AST, Viral Phase, Level of Fibrosis and Inflammation
3.7. PCM1 Genes Are Associated with Clinical Outcome in HCC
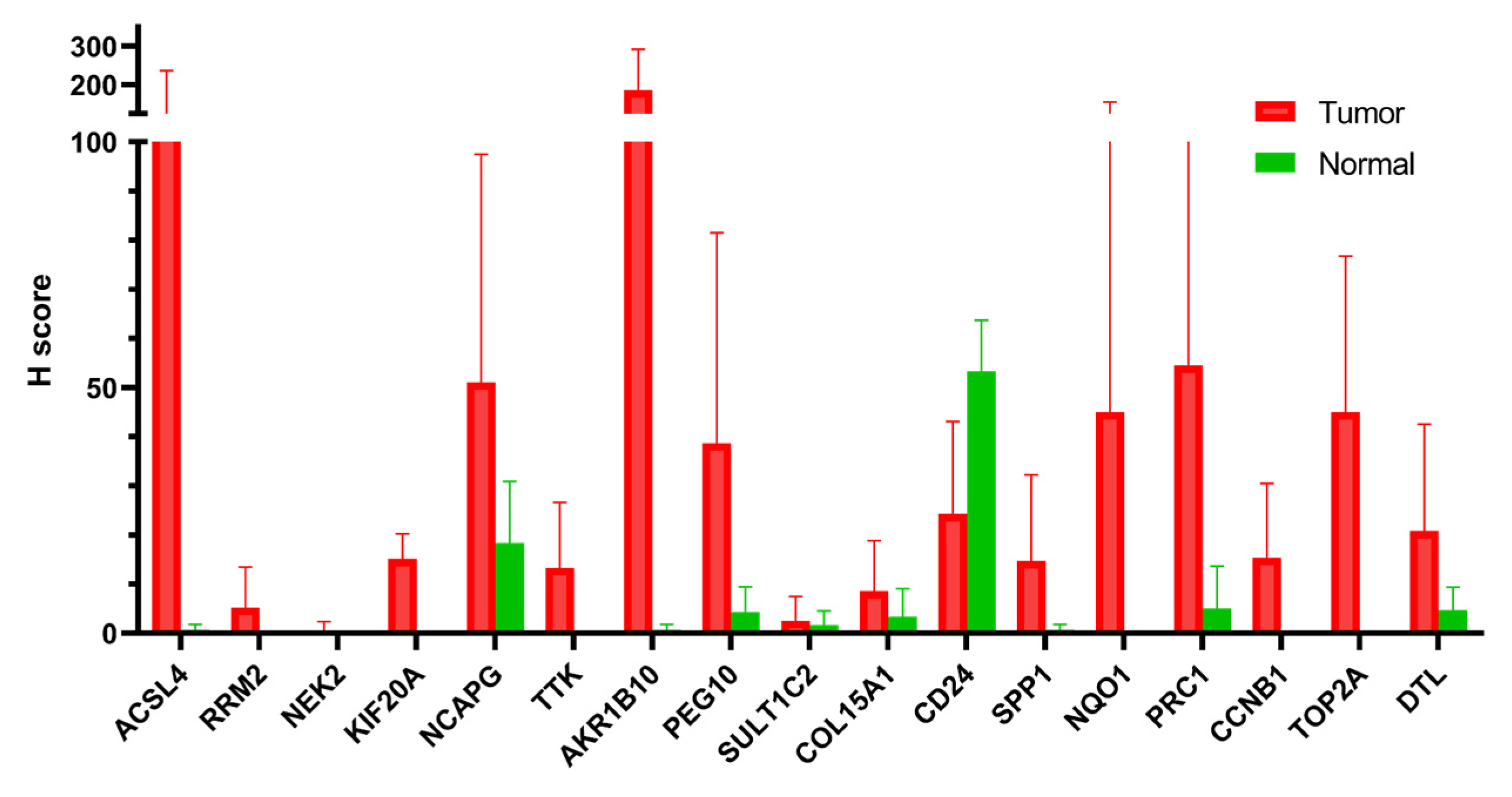
3.8. PCM1 Genes Validated by Immunohistochemistry in Hepatocellular Cancer Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 2006, 118, 3030–3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.J.; Iloeje, U.H.; Yang, H.I. Long-term outcomes in hepatitis B: The REVEAL-HBV study. Clin. Liver Dis. 2007, 11, 797–816. [Google Scholar] [CrossRef]
- World Health Organization. Global Hepatitis Report 2017; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Russo, F.P.; Rodriguez-Castro, K.; Scribano, L.; Gottardo, G.; Vanin, V.; Farinati, F. Role of antiviral therapy in the natural history of hepatitis B virus-related chronic liver disease. World J. Hepatol. 2015, 7, 1097–1104. [Google Scholar] [CrossRef]
- Terrault, N.A.; Lok, A.S.F.; McMahon, B.J.; Chang, K.M.; Hwang, J.P.; Jonas, M.M.; Brown, R.S., Jr.; Bzowej, N.H.; Wong, J.B. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018, 67, 1560–1599. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.M.; Mahtab, M.A.; Akbar, S.M.; Kamal, M.; Rahman, S. Hepatic necroinflammation and severe liver fibrosis in patients with chronic hepatitis B with undetectable HBV DNA and persistently normal alanine aminotransferase. Bangladesh Med. Res. Counc. Bull. 2014, 40, 92–96. [Google Scholar] [CrossRef]
- Lau, D.T.; Bleibel, W. Current status of antiviral therapy for hepatitis B. Ther. Adv. Gastroenterol. 2008, 1, 61–75. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Ahn, S.H. Prediction models of hepatocellular carcinoma development in chronic hepatitis B patients. World J. Gastroenterol. 2016, 22, 8314–8321. [Google Scholar] [CrossRef]
- Seo, Y.S.; Jang, B.K.; Um, S.H.; Hwang, J.S.; Han, K.H.; Kim, S.G.; Lee, K.S.; Kim, S.U.; Kim, Y.S.; Lee, J.I. Validation of risk prediction models for the development of HBV-related HCC: A retrospective multi-center 10-year follow-up cohort study. Oncotarget 2017, 8, 113213–113224. [Google Scholar] [CrossRef] [Green Version]
- Furuta, M.; Tanaka, H.; Shiraishi, Y.; Unida, T.; Imamura, M.; Fujimoto, A.; Fujita, M.; Sasaki-Oku, A.; Maejima, K.; Nakano, K.; et al. Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget 2018, 9, 25075–25088. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Dong, S.; Qiao, F.; Lu, S.; Song, Y.; Lao, Y.; Li, Y.; Zeng, T.; Hu, J.; Zhang, L.; et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene 2013, 32, 3296–3305. [Google Scholar] [CrossRef] [Green Version]
- Lamontagne, J.; Mell, J.C.; Bouchard, M.J. Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression. PLoS Pathog. 2016, 12, e1005438. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Ma, Y.; Zhang, J.; Hu, J.; Zhang, M.; Wang, Y.; Li, Y.; Wu, L.; Pan, Y.; Zhang, Y.; et al. Predictive model for inflammation grades of chronic hepatitis B: Large-scale analysis of clinical parameters and gene expressions. Liver Int. 2017, 37, 1632–1641. [Google Scholar] [CrossRef]
- Wang, M.; Gong, Q.; Zhang, J.; Chen, L.; Zhang, Z.; Lu, L.; Yu, D.; Han, Y.; Zhang, D.; Chen, P.; et al. Characterization of gene expression profiles in HBV-related liver fibrosis patients and identification of ITGBL1 as a key regulator of fibrogenesis. Sci. Rep. 2017, 7, 43446. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, F.; Zhang, X.; Yu, J.; Wang, J.; Jia, J.; Yu, X.; Shen, Z.; Yuan, Z.; Zhang, X.; et al. Differentially Expressed Intrahepatic Genes Contribute to Control of Hepatitis B Virus Replication in the Inactive Carrier Phase. J. Infect. Dis. 2018, 217, 1044–1054. [Google Scholar] [CrossRef]
- Hou, J.; Brouwer, W.P.; Kreefft, K.; Gama, L.; Price, S.L.; Janssen, H.L.A.; French, P.J.; Vanwolleghem, T.; Boonstra, A. Unique intrahepatic transcriptomics profiles discriminate the clinical phases of a chronic HBV infection. PLoS ONE 2017, 12, e0179920. [Google Scholar] [CrossRef] [Green Version]
- Roessler, S.; Jia, H.L.; Budhu, A.; Forgues, M.; Ye, Q.H.; Lee, J.S.; Thorgeirsson, S.S.; Sun, Z.; Tang, Z.Y.; Qin, L.X.; et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010, 70, 10202–10212. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.M.; Ooi, L.L.; Hui, K.M. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin. Cancer Res. 2007, 13, 6275–6283. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Kibbe, W.A.; Lin, S.M. Lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunning, M.; Lynch, A.; Eldridge, M. Illuminahumanv4.db: Illumina HumanHT12v4 Annotation Data (Chip Illuminahumanv4); R Package: Vienna, Austria, 2015. [Google Scholar]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Ponten, F.; Jirstrom, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- de Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [Green Version]
- Saldanha, A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef] [Green Version]
- Hossain, S. Visualization of Bioinformatics Data with Dash Bio. In Proceedings of the 18th Python in Science Conference, Austin, TX, USA, 8–14 July 2019. [Google Scholar]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T. A Package for Survival Analysis in R; R Package: Vienna, Austria, 2022. [Google Scholar]
- Behesti, H.; Fore, T.R.; Wu, P.; Horn, Z.; Leppert, M.; Hull, C.; Hatten, M.E. ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins. Proc. Natl. Acad. Sci. USA 2018, 115, E9717–E9726. [Google Scholar] [CrossRef] [Green Version]
- Sittler, A.; Walter, S.; Wedemeyer, N.; Hasenbank, R.; Scherzinger, E.; Eickhoff, H.; Bates, G.P.; Lehrach, H.; Wanker, E.E. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol. Cell 1998, 2, 427–436. [Google Scholar] [CrossRef]
- Liu, F.; Bao, Y.; Qiu, B.; Mao, J.; Liao, X.; Huang, H.; Zhang, A.; Zhang, G.; Qi, S.; Mei, F. Identification of Novel Cerebrospinal Fluid Biomarkers for Cognitive Decline in Aneurysmal Subarachnoid Hemorrhage: A Proteomic Approach. Front. Cell. Neurosci. 2022, 16, 861425. [Google Scholar] [CrossRef]
- Mizushima, K.; Miyamoto, Y.; Tsukahara, F.; Hirai, M.; Sakaki, Y.; Ito, T. A novel G-protein-coupled receptor gene expressed in striatum. Genomics 2000, 69, 314–321. [Google Scholar] [CrossRef]
- Cai, Y.N.; Zhou, Q.; Kong, Y.Y.; Li, M.; Viollet, B.; Xie, Y.H.; Wang, Y. LRH-1/hB1F and HNF1 synergistically up-regulate hepatitis B virus gene transcription and DNA replication. Cell Res. 2003, 13, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Miura, H.; Tomaru, Y.; Nakanishi, M.; Kondo, S.; Hayashizaki, Y.; Suzuki, M. Identification of DNA regions and a set of transcriptional regulatory factors involved in transcriptional regulation of several human liver-enriched transcription factor genes. Nucleic Acids Res. 2009, 37, 778–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaber, S.; Paugam-Burtz, C. Acute liver failure and elevated troponin-I: Controversial results and significance? Crit. Care 2013, 17, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enzan, H.; Himeno, H.; Iwamura, S.; Onishi, S.; Saibara, T.; Yamamoto, Y.; Hara, H. Alpha-smooth muscle actin-positive perisinusoidal stromal cells in human hepatocellular carcinoma. Hepatology 1994, 19, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Tyurina, D.A.; Ivanova, O.N.; Kochetkov, S.N.; Bartosch, B.; Isaguliants, M.G. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget 2017, 8, 3895–3932. [Google Scholar] [CrossRef] [Green Version]
- Dimri, M.; Satyanarayana, A. Molecular Signaling Pathways and Therapeutic Targets in Hepatocellular Carcinoma. Cancers 2020, 12, 491. [Google Scholar] [CrossRef] [Green Version]
- Boyault, S.; Rickman, D.S.; de Reynies, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Herault, A.; Saric, J.; Belghiti, J.; Franco, D.; et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Chiang, D.Y.; Villanueva, A.; Hoshida, Y.; Peix, J.; Newell, P.; Minguez, B.; LeBlanc, A.C.; Donovan, D.J.; Thung, S.N.; Sole, M.; et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008, 68, 6779–6788. [Google Scholar] [CrossRef] [Green Version]
- Hoshida, Y.; Nijman, S.M.; Kobayashi, M.; Chan, J.A.; Brunet, J.P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Chu, I.S.; Heo, J.; Calvisi, D.F.; Sun, Z.; Roskams, T.; Durnez, A.; Demetris, A.J.; Thorgeirsson, S.S. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004, 40, 667–676. [Google Scholar] [CrossRef]
- Teufel, A.; Staib, F.; Kanzler, S.; Weinmann, A.; Schulze-Bergkamen, H.; Galle, P.R. Genetics of hepatocellular carcinoma. World J. Gastroenterol. 2007, 13, 2271–2282. [Google Scholar] [CrossRef]
- Ha, H.; Debnath, B.; Neamati, N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases. Theranostics 2017, 7, 1543–1588. [Google Scholar] [CrossRef]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy--from molecular mechanisms to therapeutic benefits. Biochim. Biophys. Acta 2005, 1754, 253–262. [Google Scholar] [CrossRef]
- Chaturvedi, V.K.; Singh, A.; Dubey, S.K.; Hetta, H.F.; John, J.; Singh, M.P. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb. Pathog. 2019, 128, 184–194. [Google Scholar] [CrossRef]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Liu, Y.C.; Zou, X.B.; Chai, Y.F.; Yao, Y.M. Macrophage polarization in inflammatory diseases. Int. J. Biol. Sci. 2014, 10, 520–529. [Google Scholar] [CrossRef] [Green Version]
- Lawand, M.; Dechanet-Merville, J.; Dieu-Nosjean, M.C. Key Features of Gamma-Delta T-Cell Subsets in Human Diseases and Their Immunotherapeutic Implications. Front. Immunol. 2017, 8, 761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajoriya, N.; Fergusson, J.R.; Leithead, J.A.; Klenerman, P. Gamma Delta T-lymphocytes in Hepatitis C and Chronic Liver Disease. Front. Immunol. 2014, 5, 400. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Han, J.; Wu, H. Regulatory T-cells promote hepatitis B virus infection and hepatocellular carcinoma progression. Chronic Dis. Transl. Med. 2016, 2, 67–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.L.; Kao, J.H. New perspectives of biomarkers for the management of chronic hepatitis B. Clin. Mol. Hepatol. 2016, 22, 423–431. [Google Scholar] [CrossRef] [Green Version]
- Niu, B.; Hann, H.W.L. Hepatitis B Virus–Related Hepatocellular Carcinoma: Carcinogenesis, Prevention, and Treatment; Books on Demand: Norderstedt, Germany, 2017. [Google Scholar]
- European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–398. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.T. Immune tolerant hepatitis B: A clinical dilemma. Gastroenterol. Hepatol. 2011, 7, 511–516. [Google Scholar]
- Scheuer, P.J. Classification of chronic viral hepatitis: A need for reassessment. J. Hepatol. 1991, 13, 372–374. [Google Scholar] [CrossRef]
- Karin, M. The IkappaB kinase—A bridge between inflammation and cancer. Cell Res. 2008, 18, 334–342. [Google Scholar] [CrossRef]
- Chung, W.; Kim, M.; de la Monte, S.; Longato, L.; Carlson, R.; Slagle, B.L.; Dong, X.; Wands, J.R. Activation of signal transduction pathways during hepatic oncogenesis. Cancer Lett. 2016, 370, 1–9. [Google Scholar] [CrossRef]
- Yang, H.I.; Yuen, M.F.; Chan, H.L.; Han, K.H.; Chen, P.J.; Kim, D.Y.; Ahn, S.H.; Chen, C.J.; Wong, V.W.; Seto, W.K.; et al. Risk estimation for hepatocellular carcinoma in chronic hepatitis B (REACH-B): Development and validation of a predictive score. Lancet Oncol. 2011, 12, 568–574. [Google Scholar] [CrossRef]
- Papatheodoridis, G.; Dalekos, G.; Sypsa, V.; Yurdaydin, C.; Buti, M.; Goulis, J.; Calleja, J.L.; Chi, H.; Manolakopoulos, S.; Mangia, G.; et al. PAGE-B predicts the risk of developing hepatocellular carcinoma in Caucasians with chronic hepatitis B on 5-year antiviral therapy. J. Hepatol. 2016, 64, 800–806. [Google Scholar] [CrossRef]
- Yuen, M.F.; Tanaka, Y.; Fong, D.Y.; Fung, J.; Wong, D.K.; Yuen, J.C.; But, D.Y.; Chan, A.O.; Wong, B.C.; Mizokami, M.; et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J. Hepatol. 2009, 50, 80–88. [Google Scholar] [CrossRef]
- Khalaf, A.M.; Fuentes, D.; Morshid, A.I.; Burke, M.R.; Kaseb, A.O.; Hassan, M.; Hazle, J.D.; Elsayes, K.M. Role of Wnt/beta-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J. Hepatocell. Carcinoma 2018, 5, 61–73. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.H.; Shi, C.H. Molecular characteristics and stages of chronic hepatitis B virus infection. World J. Gastroenterol. 2009, 15, 3099–3105. [Google Scholar] [CrossRef]
- Sharma, S.K.; Saini, N.; Chwla, Y. Hepatitis B virus: Inactive carriers. Virol. J. 2005, 2, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Z.; Hou, X.; Liu, W.; Han, Z.; Wei, L. Macrophages and hepatocellular carcinoma. Cell Biosci. 2019, 9, 79. [Google Scholar] [CrossRef] [Green Version]
- Bernhard, S.; Hug, S.; Stratmann, A.E.P.; Erber, M.; Vidoni, L.; Knapp, C.L.; Thomass, B.D.; Fauler, M.; Nilsson, B.; Nilsson Ekdahl, K.; et al. Interleukin 8 Elicits Rapid Physiological Changes in Neutrophils That Are Altered by Inflammatory Conditions. J. Innate Immun. 2021, 13, 225–241. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Huh, K.W.; Siddiqui, A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol. Cell. Biol. 2001, 21, 7721–7730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.X.; Huang, C.J.; Yang, Z.G. Impact of hepatitis B virus infection on hepatic metabolic signaling pathway. World J. Gastroenterol. 2016, 22, 8161–8167. [Google Scholar] [CrossRef]
- Bertoletti, A.; Kennedy, P.T. The immune tolerant phase of chronic HBV infection: New perspectives on an old concept. Cell. Mol. Immunol. 2015, 12, 258–263. [Google Scholar] [CrossRef] [Green Version]
- Hou, W.; Wu, X. Diverse Functions of gammadelta T Cells in the Progression of Hepatitis B Virus and Hepatitis C Virus Infection. Front. Immunol. 2020, 11, 619872. [Google Scholar] [CrossRef]
- Edwards, S.C.; Sutton, C.E.; Ladell, K.; Grant, E.J.; McLaren, J.E.; Roche, F.; Dash, P.; Apiwattanakul, N.; Awad, W.; Miners, K.L.; et al. A population of proinflammatory T cells coexpresses alphabeta and gammadelta T cell receptors in mice and humans. J. Exp. Med. 2020, 217, e20190834. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Zhang, D.; Zhen, W.; Shi, Q.; Liu, Y.; Ling, N.; Peng, M.; Tang, K.; Hu, P.; Hu, H.; et al. Characteristics of circulating T cell receptor gamma-delta T cells from individuals chronically infected with hepatitis B virus (HBV): An association between V(delta)2 subtype and chronic HBV infection. J. Infect. Dis. 2008, 198, 1643–1650. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Hu, P.; Ling, N.; Peng, H.; Lei, Y.; Hu, H.; Zhang, D.; Ren, H. Enhanced functions of peripheral gammadelta T cells in chronic hepatitis B infection during interferon alpha treatment in vivo and in vitro. PLoS ONE 2015, 10, e0120086. [Google Scholar] [CrossRef]
- Matkowskyj, K.A.; Bai, H.; Liao, J.; Zhang, W.; Li, H.; Rao, S.; Omary, R.; Yang, G.Y. Aldoketoreductase family 1B10 (AKR1B10) as a biomarker to distinguish hepatocellular carcinoma from benign liver lesions. Hum. Pathol. 2014, 45, 834–843. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Yan, R.; Luo, D.; Watabe, K.; Liao, D.F.; Cao, D. Aldo-keto reductase family 1 member B10 promotes cell survival by regulating lipid synthesis and eliminating carbonyls. J. Biol. Chem. 2009, 284, 26742–26748. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Liao, W.; Yao, W.; Zhu, R.; Li, Y.; He, S. Aldo-keto Reductase Family 1 Member B 10 Mediates Liver Cancer Cell Proliferation through Sphingosine-1-Phosphate. Sci. Rep. 2016, 6, 22746. [Google Scholar] [CrossRef] [Green Version]
- Petrizzo, A.; Caruso, F.P.; Tagliamonte, M.; Tornesello, M.L.; Ceccarelli, M.; Costa, V.; Aprile, M.; Esposito, R.; Ciliberto, G.; Buonaguro, F.M.; et al. Identification and Validation of HCC-specific Gene Transcriptional Signature for Tumor Antigen Discovery. Sci. Rep. 2016, 6, 29258. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Genda, T.; Ichida, T.; Murata, A.; Tsuzura, H.; Narita, Y.; Kanemitsu, Y.; Ishikawa, S.; Kikuchi, T.; Mori, M.; et al. Impact of aldo-keto reductase family 1 member B10 on the risk of hepatitis C virus-related hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2016, 31, 1315–1322. [Google Scholar] [CrossRef]
- Mori, M.; Genda, T.; Ichida, T.; Murata, A.; Kamei, M.; Tsuzura, H.; Sato, S.; Narita, Y.; Kanemitsu, Y.; Ishikawa, S.; et al. Aldo-keto reductase family 1 member B10 is associated with hepatitis B virus-related hepatocellular carcinoma risk. Hepatol. Res. 2017, 47, E85–E93. [Google Scholar] [CrossRef]
- Han, C.; Gao, L.; Bai, H.; Dou, X. Identification of a role for serum aldo-keto reductase family 1 member B10 in early detection of hepatocellular carcinoma. Oncol. Lett. 2018, 16, 7123–7130. [Google Scholar] [CrossRef] [Green Version]
- Maloberti, P.; Castilla, R.; Castillo, F.; Cornejo Maciel, F.; Mendez, C.F.; Paz, C.; Podesta, E.J. Silencing the expression of mitochondrial acyl-CoA thioesterase I and acyl-CoA synthetase 4 inhibits hormone-induced steroidogenesis. FEBS J. 2005, 272, 1804–1814. [Google Scholar] [CrossRef]
- Macias-Rodriguez, R.U.; Inzaugarat, M.E.; Ruiz-Margain, A.; Nelson, L.J.; Trautwein, C.; Cubero, F.J. Reclassifying Hepatic Cell Death during Liver Damage: Ferroptosis—A Novel Form of Non-Apoptotic Cell Death? Int. J. Mol. Sci. 2020, 21, 1651. [Google Scholar] [CrossRef] [Green Version]
| PCM1-U | ALT (IU/L) | |||||
| AST | ≤40 | >40 | HBVDNA | |||
| (IU/L) | GSE83148 | GSE84044 | GSE83148 | GSE84044 | (Copies/mL) | |
| ≤35 | 0 * | 0 | 0 | 0 | ≤1 × 106 | |
| >35 | 0 | 0 | 14 | 15 | ≤1 × 106 | |
| ≤35 | 0 | 0 | 0 | 1 | >1 × 106 | |
| >35 | 0 | 1 | 15 | 16 | >1 × 106 | |
| PCM1-D | ALT (IU/L) | |||||
| AST | ≤40 | >40 | HBVDNA | |||
| GSE83148 | GSE84044 | GSE83148 | GSE84044 | (Copies/mL) | ||
| ≤35 | 8 * | 14 | 1 | 4 | ≤1 × 106 | |
| >35 | 2 | 1 | 7 | 11 | ≤1 × 106 | |
| ≤35 | 13 | 16 | 4 | 3 | >1 × 106 | |
| >35 | 2 | 2 | 18 | 14 | >1 × 106 | |
| GSE83148 | ||||||
| PCM1-U | PCM1-D | X2 | p Value * | |||
| Clinical Data | n ** | % ** | n ** | % ** | ||
| ALT (IU/L): | ||||||
| ≤40 | 1 | 3.30% | 37 | 49.30% | 19.635 a | <0.001 |
| >40 | 29 | 96.70% | 38 | 50.70% | ||
| AST (IU/L): | ||||||
| ≤35 | 1 | 3.30% | 41 | 54.70% | 23.528 b | <0.001 |
| >35 | 29 | 96.70% | 34 | 45.30% | ||
| HBV DNA (copies/mL): | ||||||
| ≤1 × 106 >1 × 106 | 14 16 | 46.70% 53.30% | 22 38 | 36.70% 63.30% | 0.833 c | 0.494 |
| GSE84044 | ||||||
| PCM1-U | PCM1-D | X2 | p Value * | |||
| Clinical Data | n ** | % ** | n ** | % ** | ||
| ALT (IU/L): | ||||||
| ≤40 | 1 | 3.00% | 39 | 53.40% | 24.564 d | <0.001 |
| >40 | 32 | 97.00% | 34 | 46.60% | ||
| AST (IU/L): | ||||||
| ≤35 | 1 | 3.00% | 43 | 59.70% | 29.874 e | <0.001 |
| >35 | 32 | 97.00% | 29 | 40.30% | ||
| HBV DNA (copies/mL): | ||||||
| ≤1 × 106 >1 × 106 | 16 19 | 45.70% 54.30% | 37 42 | 46.80% 53.20% | 0.012 f | 1.000 |
| Viral Phase | PCM1-U | PCM1-D | Chi-Square | ||
| n * | % * | n * | % * | X2 = 16.8135, p < 0.001 | |
| Immune Tolerance | 0 | 0.00% | 22 | 40.70% | |
| Immune Clearance | 25 | 86.20% | 25 | 46.30% | |
| Inactive Carrier | 4 | 13.80% | 7 | 13.00% | |
| Total | 29 | 100% | 54 | 100% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaoglu, D.A.; Uner, M.; Simsek, C.; Gure, A.O.; Demirkol-Canli, S. Transcriptomic Analysis of Hepatitis B Infected Liver for Prediction of Hepatocellular Carcinoma. Biology 2023, 12, 188. https://doi.org/10.3390/biology12020188
Karaoglu DA, Uner M, Simsek C, Gure AO, Demirkol-Canli S. Transcriptomic Analysis of Hepatitis B Infected Liver for Prediction of Hepatocellular Carcinoma. Biology. 2023; 12(2):188. https://doi.org/10.3390/biology12020188
Chicago/Turabian StyleKaraoglu, Diren Arda, Meral Uner, Cem Simsek, Ali Osmay Gure, and Secil Demirkol-Canli. 2023. "Transcriptomic Analysis of Hepatitis B Infected Liver for Prediction of Hepatocellular Carcinoma" Biology 12, no. 2: 188. https://doi.org/10.3390/biology12020188





