Overview of Yersinia pestis Metallophores: Yersiniabactin and Yersinopine
Abstract
:Simple Summary
Abstract
1. Introduction
2. Yersiniabactin
2.1. Yersiniabactin Biosynthesis
2.2. The Export of Ybt
2.3. Iron Chelation by Ybt
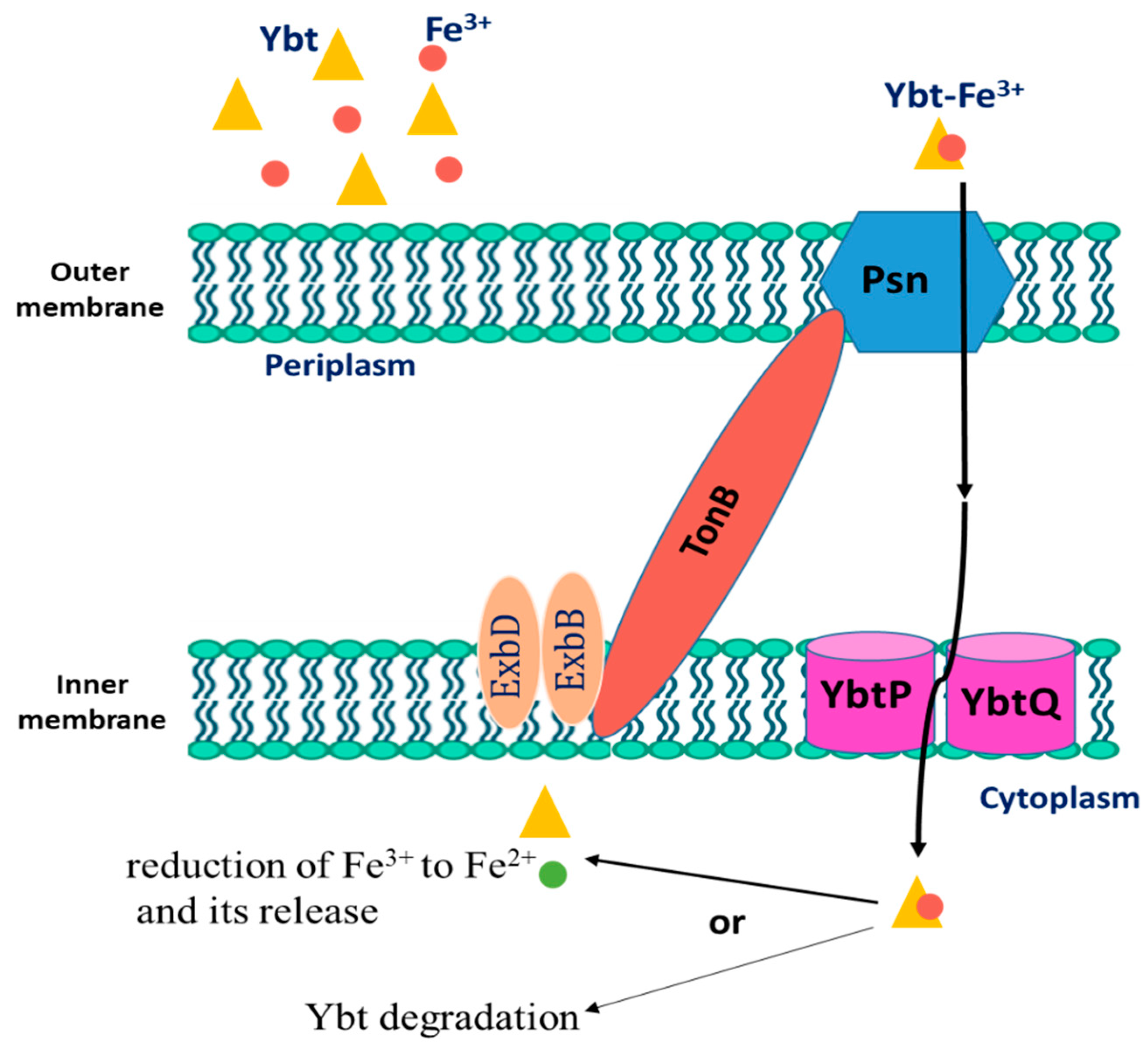
2.4. The Intake of Iron-Loaded Ybt
2.5. Genetic Regulation of Yersiniabactin (Ybt)
2.6. The Role of Yersiniabactin in Overcoming Zinc Restriction
3. Yersinopine
4. Aerobactin
5. The Evolution of Y. pestis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, R.; Atkinson, S.; Chen, Z.; Cui, Y.; Du, Z.; Han, Y.; Sebbane, F.; Slavin, P.; Song, Y.; Yan, Y.; et al. Yersinia pestis and Plague: Some Knowns and Unknowns. Zoonoses 2023, 3. [Google Scholar] [CrossRef]
- Eisen, R.J.; Bearden, S.W.; Wilder, A.P.; Montenieri, J.A.; Antolin, M.F.; Gage, K.L. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc. Natl. Acad. Sci. USA 2006, 103, 15380–15385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinnebusch, B.J.; Jarrett, C.O.; Bland, D.M. “Fleaing” the plague: Adaptations of Yersinia pestis to its insect vector that lead to transmission. Annu. Rev. Microbiol. 2017, 71, 215–232. [Google Scholar] [CrossRef]
- Lathem, W.W.; Crosby, S.D.; Miller, V.L.; Goldman, W.E. Progression of primary pneumonic plague: A mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. USA 2005, 102, 17786–17791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pechous, R.D.; Sivaraman, V.; Stasulli, N.M.; Goldman, W.E. Pneumonic plague: The darker side of Yersinia pestis. Trends Microbiol. 2016, 24, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Dean, K.R.; Krauer, F.; Walløe, L.; Lingjærde, O.C.; Bramanti, B.; Stenseth, N.C.; Schmid, B.V. Human ectoparasites and the spread of plague in Europe during the Second Pandemic. Proc. Natl. Acad. Sci. USA 2018, 115, 1304–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demeure, C.E.; Dussurget, O.; Fiol, G.M.; Le Guern, A.-S.; Savin, C.; Pizarro-Cerdá, J. Yersinia pestis and plague: An updated view on evolution, virulence determinants, immune subversion, vaccination, and diagnostics. Genes Immun. 2019, 20, 357–370. [Google Scholar] [CrossRef] [Green Version]
- Montminy, S.W.; Khan, N.; McGrath, S.; Walkowicz, M.J.; Sharp, F.; Conlon, J.E.; Fukase, K.; Kusumoto, S.; Sweet, C.; Miyake, K.; et al. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat. Immunol. 2006, 7, 1066–1073. [Google Scholar] [CrossRef]
- Mordechai, L.; Eisenberg, M.; Newfield, T.P.; Izdebski, A.; Kay, J.E.; Poinar, H. The Justinianic Plague: An inconsequential pandemic? Proc. Natl. Acad. Sci. USA 2019, 116, 25546–25554. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.; Spyrou, M.A.; Scheib, C.L.; Neumann, G.U.; Kröpelin, A.; Haas-Gebhard, B.; Päffgen, B.; Haberstroh, J.; Ribera, I.; Lacomba, A.; et al. Ancient Yersinia pestis genomes from across Western Europe reveal early diversification during the First Pandemic (541–750). Proc. Natl. Acad. Sci. USA 2019, 116, 12363–12372. [Google Scholar] [CrossRef] [Green Version]
- Namouchi, A.; Guellil, M.; Kersten, O.; Hänsch, S.; Ottoni, C.; Schmid, B.V.; Pacciani, E.; Quaglia, L.; Vermunt, M.; Bauer, E.L.; et al. Integrative approach using Yersinia pestis genomes to revisit the historical landscape of plague during the Medieval Period. Proc. Natl. Acad. Sci. USA 2018, 115, 11790–11797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, R.J.; Miller, V.L. A deadly path: Bacterial spread during bubonic plague. Trends Microbiol. 2016, 24, 239–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbaji, A.; Kharabsheh, S.; Al-Azab, S.; Al-Kayed, M.; Amr, Z.S.; Abu Baker, M.; Chu, M.C. A 12-case outbreak of pharyngeal plague following the consumption of camel meat. Ann. Trop. Med. Parasitol. 2005, 99, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Randremanana, R.; Andrianaivoarimanana, V.; Nikolay, B.; Ramasindrazana, B.; Paireau, J.; ten Bosch, Q.A.; Rakotondramanga, J.M.; Rahajandraibe, S.; Rahelinirina, S.; Rakotomanana, F.; et al. Epidemiological characteristics of an urban plague epidemic in Madagascar, August–November, 2017: An outbreak report. Lancet Infect. Dis. 2019, 19, 537–545. [Google Scholar] [CrossRef] [Green Version]
- Guiyoule, A.; Gerbaud, G.; Buchrieser, C.; Galimand, M.; Rahalison, L.; Chanteau, S.; Courvalin, P.; Carniel, E. Transferable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis. Emerg. Infect. Dis. 2001, 7, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galimand, M.; Carniel, E.; Courvalin, P. Resistance of Yersinia pestis to antimicrobial agents. Antimicrob. Agents Chemother. 2006, 50, 3233–3236. [Google Scholar] [CrossRef] [Green Version]
- Cornelius, C.A.; Quenee, L.E.; Overheim, K.A.; Koster, F.; Brasel, T.L.; Elli, D.; Ciletti, N.A.; Schneewind, O. Immunization with recombinant V10 protects cynomolgus macaques from lethal pneumonic plague. Infect. Immun. 2008, 76, 5588–5597. [Google Scholar] [CrossRef] [Green Version]
- Becker, K.W.; Skaar, E.P. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 2014, 38, 1235–1249. [Google Scholar] [CrossRef] [Green Version]
- Lopez, C.A.; Skaar, E.P. The impact of dietary transition metals on host-bacterial interactions. Cell Host Microbe 2018, 23, 737–748. [Google Scholar] [CrossRef] [Green Version]
- Skaar, E.P.; Raffatellu, M. Metals in infectious diseases and nutritional immunity. Metallomics 2015, 7, 926–928. [Google Scholar] [CrossRef]
- Lonergan, Z.R.; Skaar, E.P. Nutrient zinc at the host-pathogen interface. Trends Biochem. Sci. 2019, 44, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Palmer, L.D.; Skaar, E.P. Transition Metals and Virulence in Bacteria. Annu. Rev. Genet. 2016, 50, 67–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps: Is immunity the second function of chromatin? J. Cell Biol. 2012, 198, 773–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohnle, P.G.; Hunter, M.J.; Hahn, B.; Chazin, W.J. Zinc-reversible antimicrobial activity of recombinant calprotectin (migration inhibitory factor-related proteins 8 and 14). J. Infect. Dis. 2000, 182, 1272–1275. [Google Scholar] [CrossRef] [PubMed]
- Kelliher, J.L.; Kehl-Fie, T.E. Competition for manganese at the host-pathogen interface. Prog. Mol. Biol. Transl. Sci. 2016, 142, 1–25. [Google Scholar] [PubMed]
- Damo, S.M.; Kehl-Fie, T.E.; Sugitani, N.; Holt, M.E.; Rathi, S.; Murphy, W.J.; Zhang, Y.; Betz, C.; Hench, L.; Fritz, G.; et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. USA 2013, 110, 3841–3846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehl-Fie, T.E.; Chitayat, S.; Hood, M.I.; Damo, S.; Restrepo, N.; Garcia, C.; Munro, K.A.; Chazin, W.J.; Skaar, E.P. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 2011, 10, 158–164. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.Z.; Jellbauer, S.; Poe, A.J.; Ton, V.; Pesciaroli, M.; Kehl-Fie, T.E.; Restrepo, N.A.; Hosking, M.P.; Edwards, R.A.; Battistoni, A.; et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 2012, 11, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Aisen, P.; Enns, C.; Wessling-Resnick, M. Chemistry and biology of eukaryotic iron metabolism. Int. J. Biochem. Cell Biol. 2001, 33, 940–959. [Google Scholar] [CrossRef]
- Braun, V.; Hantke, K. Recent insights into iron import by bacteria. Curr. Opin. Chem. Biol. 2011, 15, 328–334. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodríguez Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masson, P.L.; Heremans, J.F.; Schonne, E. Lactoferrin, an iron-binding protein in neutrophilic leukocytes. J. Exp. Med. 1969, 130, 643–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakin, A.; Schneider, L.; Podladchikova, O. Hunger for iron: The alternative siderophore iron scavenging systems in highly virulent Yersinia. Front. Cell. Infect. Microbiol. 2012, 2, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schalk, I.J.; Hannauer, M.; Braud, A. New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef] [PubMed]
- Lhospice, S.; Gomez, N.O.; Ouerdane, L.; Brutesco, C.; Ghssein, G.; Hajjar, C.; Liratni, A.; Wang, S.; Richaud, P.; Bleves, S.; et al. Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 2017, 7, 17132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelissen, C.N.; Hollander, A. TonB-Dependent Transporters Expressed by Neisseria gonorrhoeae. Front. Microbiol. 2011, 2, 117. [Google Scholar] [CrossRef] [Green Version]
- Dale, S.E.; Doherty-Kirby, A.; Lajoie, G.; Heinrichs, D.E. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: Identification and characterization of genes involved in production of a siderophore. Infect. Immun. 2004, 72, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Ghssein, G.; Matar, S.F. Chelating Mechanisms of Transition Metals by Bacterial Metallophores “Pseudopaline and Staphylopine”: A Quantum Chemical Assessment. Computation 2018, 6, 56. [Google Scholar] [CrossRef] [Green Version]
- Mastropasqua, M.C.; D′Orazio, M.; Cerasi, M.; Pacello, F.; Gismondi, A.; Canini, A.; Canuti, L.; Consalvo, A.; Ciavardelli, D.; Chirullo, B.; et al. Growth of Pseudomonas aeruginosa in zinc poor environments is promoted by a nicotianamine-related metallophore. Mol. Microbiol. 2017, 106, 543–561. [Google Scholar] [CrossRef] [Green Version]
- Perry, R.D.; Balbo, P.B.; Jones, H.A.; Fetherston, J.D.; DeMoll, E. Yersiniabactin from Yersinia pestis: Biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 1999, 145, 1181–1190. [Google Scholar] [CrossRef] [Green Version]
- McFarlane, J.S.; Davis, C.L.; Lamb, A.L. Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase. J. Biol. Chem. 2018, 293, 8009–8019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakin, A.; Saken, E.; Harmsen, D.; Heesemann, J. The pesticin receptor of Yersinia enterocolitica: A novel virulence factor with dual function. Mol. Microbiol. 1994, 13, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Fetherston, J.D.; Kirillina, O.; Bobrov, A.G.; Paulley, J.T.; Perry, R.D. The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun. 2010, 70, 2045–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee-Lewis, H.; Anderson, D.M. Absence of inflammation and pneumonia during infection with nonpigmented Yersinia pestis reveals a new role for the pgm locus in pathogenesis. Infect. Immun. 2009, 78, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, R.D.; Fetherston, J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–817. [Google Scholar] [CrossRef] [Green Version]
- Chambers, C.E.; McIntyre, D.D.; Mouck, M.; Sokol, P.A. Physical and structural characterization of yersiniophore, a siderophore produced by clinical isolates of Yersinia enterocolitica. Biometals 1996, 9, 157–167. [Google Scholar] [CrossRef]
- Bearden, S.W.; Fetherston, J.D.; Perry, R.D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 1997, 65, 1659–1668. [Google Scholar] [CrossRef] [Green Version]
- Gehring, A.M.; DeMoll, E.; Fetherston, J.D.; Mori, I.; Mayhew, G.F.; Blattner, F.R.; Walsh, C.T.; Perry, R.D. Iron acquisition in plague: Modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 1998, 5, 573–586. [Google Scholar] [CrossRef] [Green Version]
- Bobrov, A.G.; Geoffroy, V.A.; Perry, R.D. Yersiniabactin production requires the thioesterase domain of HMWP2 and YbtD, a putative phosphopantetheinylate transferase. Infect. Immun. 2002, 70, 4204–4214. [Google Scholar] [CrossRef] [Green Version]
- Geoffroy, V.A.; Fetherston, J.D.; Perry, R.D. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 2000, 68, 4452–4461. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.C.; Fetherston, J.D.; Pickett, C.L.; Bobrov, A.G.; Weaver, R.H.; DeMoll, E.; Perry, R.D. Reduced synthesis of the Ybt siderophore or production of aberrant Ybt-like molecules activates transcription of yersiniabactin genes in Yersinia pestis. Microbiology 2010, 156, 2226–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furrer, J.L.; Sanders, D.N.; Hook-Barnard, I.G.; McIntosh, M.A. Export of the siderophore enterobactin in Escherichia coli: Involvement of a 43 kDa membrane exporter. Mol. Microbiol. 2002, 44, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Brickman, T.J.; Armstrong, S.K. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 2005, 187, 3650–3661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drechsel, H.; Stephan, H.; Lotz, R.; Haag, H.; Zähner, H.; Hantke, K.; Jung, G. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann. 1995, 10, 1727–1733. [Google Scholar] [CrossRef]
- Fetherston, J.D.; Bertolino, V.J.; Perry, R.D. YbtP and YbtQ: Two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 1999, 32, 289–299. [Google Scholar] [CrossRef]
- Buchanan, S.K. Type I secretion and multidrug efflux: Transport through the TolC channel-tunnel. Trends Biochem. Sci. 2001, 28, 3–6. [Google Scholar] [CrossRef]
- Fetherston, J.D.; Lillard, J.W.; Perry, R.D. Analysis of the pesticin receptor from Yersinia pestis: Role in iron-deficient growth and possible regulation by its siderophore. J. Bacteriol. 1995, 177, 1824–1833. [Google Scholar] [CrossRef] [Green Version]
- Perry, R.D.; Shah, J.; Bearden, S.W.; Thompson, J.M.; Fetherston, J.D. Yersinia pestis TonB: Role in iron, heme and hemoprotein utilization. Infect. Immun. 2003, 71, 4159–4162. [Google Scholar] [CrossRef] [Green Version]
- Lesic, B.; Carniel, E. Yersinia Molecular and Cellular Biology. In Horizon Bioscience; Carniel, E., Hinnebusch, B.J., Eds.; UCL Press: Norfolk, UK, 2004; pp. 285–306. [Google Scholar]
- Leal-Balbino, T.C.; Leal, N.C.; Nascimento, M.G.M.D.; Oliveira, M.B.M.D.; Balbino, V.D.Q.; Almeida, A.M.P.D. The pgm locus and pigmentation phenotype in Yersinia pestis. Genet. Mol. Biol. 2006, 29, 126–131. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Zhou, D.; Li, Y.; Guo, Z.; Han, Y.; Song, Y.; Zhai, J.; Du, Z.; Wang, X.; Lu, J.; et al. The ironresponsive Fur regulon in Yersinia pestis. J. Bacteriol. 2008, 190, 3063–3075. [Google Scholar] [CrossRef] [Green Version]
- Price, S.L.; Vadyvaloo, V.; DeMarco, J.K.; Brady, A.; Gray, P.A.; Kehl-Fie, T.E.; Garneau-Tsodikova, S.; Perry, R.D.; Lawrenz, M.B. Yersiniabactin contributes to overcoming zinc restriction during Yersinia pestis infection of mammalian and insect hosts. Proc. Natl. Acad. Sci. USA 2021, 118, 2104073118. [Google Scholar] [CrossRef] [PubMed]
- Bobrov, A.G.; Kirillina, O.; Fetherston, J.D.; Miller, M.C.; Burlison, J.A.; Perry, R.D. The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol. Microbiol. 2014, 93, 759–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobrov, A.G.; Kirillina, O.; Fosso, M.Y.; Fetherston, J.D.; Miller, M.C.; VanCleave, T.T.; Burlison, J.A.; Arnold, W.K.; Lawrenz, M.B.; Garneau-Tsodikova, S.; et al. Zinc transporters YbtX and ZnuABC are required for the virulence of Yersinia pestis in bubonic and pneumonic plague in mice. Metallomics 2017, 9, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Reichman, S.M.; Parker, D.R. Revisiting the metal-binding chemistry of nicotianamine and 2′-deoxymugineic acid. Implications for iron nutrition in strategy II plants. Plant Physiol. 2002, 129, 1435–1438. [Google Scholar] [CrossRef] [Green Version]
- Trampczynska, A.; Böttcher, C.; Clemens, S. The transition metal chelator nicotianamine is synthesized by filamentous fungi. FEBS Lett. 2006, 580, 3173–3178. [Google Scholar] [CrossRef] [Green Version]
- Burkhead, J.L.; Gogolin Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review. Biology 2022, 11, 1525. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. A Review of Pseudomonas aeruginosa Metallophores: Pyoverdine, Pyochelin and Pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Ghssein, G.; Brutesco, C.; Ouerdane, L.; Fojcik, C.; Izaute, A.; Wang, S.; Hajjar, C.; Lobinski, R.; Lemaire, D.; Richaud, P.; et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 2016, 352, 1105–1109. [Google Scholar] [CrossRef]
- McFarlane, J.S.; Lamb, A.L. Biosynthesis of an opine metallophore by Pseudomonas aeruginosa. Biochemistry 2017, 56, 5967–5971. [Google Scholar] [CrossRef]
- Laffont, C.; Brutesco, C.; Hajjar, C.; Cullia, G.; Fanelli, R.; Ouerdane, L.; Cavelier, F.; Arnoux, P. Simple rules govern the diversity of bacterial nicotianamine-like metallophores. Biochem. J. 2019, 476, 2221–2233. [Google Scholar] [CrossRef]
- Remy, L.; Carrière, M.; Derré-Bobillot, A.; Martini, C.; Sanguinetti, M.; Borezée-Durant, E. The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc depleted conditions and contributes to virulence: Nickel and cobalt uptake in Staphylococcus aureus. Mol. Microbiol. 2013, 87, 730–743. [Google Scholar] [CrossRef] [PubMed]
- Bielecki, P.; Puchałka, J.; Wos-Oxley, M.L.; Loessner, H.; Glik, J.; Kawecki, M.; Nowak, M.; Tümmler, B.; Weiss, S.; dos Santos, V.A.P.M. In-Vivo Expression Profiling of Pseudomonas aeruginosa Infections Reveals Niche-Specific and Strain-Independent Transcriptional Programs. PLoS ONE 2011, 6, e24235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gi, M.; Lee, K.M.; Kim, S.C.; Yoon, J.H.; Yoon, S.S.; Choi, J.Y. A novel siderophore system is essential for the growth of Pseudomonas aeruginosa in airway mucus. Sci. Rep. 2015, 5, 14644. [Google Scholar] [CrossRef]
- Ding, Y.; Fu, Y.; Lee, J.C.; Hooper, D.C. Staphylococcus aureus NorD, a Putative Efflux Pump Coregulated with the Opp1 Oligopeptide Permease, Contributes Selectively to Fitness In Vivo. J. Bacteriol. 2012, 194, 6586–6593. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Pan, D.; Li, M.; Wang, Y.; Song, L.; Yu, D.; Zuo, Y.; Wang, K.; Liu, Y.; Wei, Z.; et al. Aerobactin-Mediated Iron Acquisition Enhances Biofilm Formation, Oxidative Stress Resistance, and Virulence of Yersinia pseudotuberculosis. Front. Microbiol. 2021, 15, 699913. [Google Scholar] [CrossRef] [PubMed]
- Forman, S.; Nagiec, M.J.; Abney, J.; Perry, R.D.; Fetherston, J.D. Analysis of the aerobactin and ferric hydroxamate uptake systems of Yersinia pestis. Microbiology 2007, 153, 2332–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkhill, J.; Wren, B.W.; Thomson, N.R.; Titball, R.W.; Holden, M.T.G.; Prentice, M.B.; Sebaihia, M.; James, K.D.; Churcher, C.; Mungall, K.L.; et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 2001, 413, 523–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achtman, M.; Zurth, K.; Morelli, G.; Torrea, G.; Guiyoule, A.; Carniel, E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 1999, 96, 14043–14048. [Google Scholar] [CrossRef] [Green Version]
- Cornelis, G.R.; Boland, A.; Boyd, A.P.; Geuijen, C.; Iriarte, M.; Neyt, C.; Sory, M.P.; Stainier, I. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 1998, 62, 1315–1352. [Google Scholar] [CrossRef] [Green Version]
- Lathem, W.W.; Price, P.A.; Miller, V.L.; Goldman, W.E. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 2007, 315, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Sebbane, F.; Jarrett, C.O.; Gardner, D.; Long, D.; Hinnebusch, B.J. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. USA 2006, 103, 5526–5530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcnally, A.; Thomson, N.R.; Reuter, S.; Wren, B.W. ‘Add, stir and reduce’: Yersinia spp. as model bacteria for pathogen evolution. Nat. Rev. Microbiol. 2016, 14, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Chain, P.S.; Hu, P.; Malfatti, S.A.; Radnedge, L.; Larimer, F.; Vergez, L.M.; Worsham, P.; Chu, M.C.; Andersen, G.L. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: Evidence of gene reduction in an emerging pathogen. J. Bacteriol. 2006, 188, 4453–4463. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wang, T.; Liu, Z.; Ke, Y.; Li, R.; Chen, H.; You, Y.; Wu, G.; Cao, S.; Du, Z.; et al. Single-cell transcriptomics of immune cells in lymph nodes reveals their composition and alterations in functional dynamics during the early stages of bubonic plague. Sci. China Life Sci. 2023, 66, 110–126. [Google Scholar] [CrossRef]
- Heroven, A.K.; Dersch, P. Coregulation of host-adapted metabolism and virulence by pathogenic yersiniae. Front. Cell. Infect. Microbiol. 2014, 4, 146. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaaban, T.; Mohsen, Y.; Ezzeddine, Z.; Ghssein, G. Overview of Yersinia pestis Metallophores: Yersiniabactin and Yersinopine. Biology 2023, 12, 598. https://doi.org/10.3390/biology12040598
Chaaban T, Mohsen Y, Ezzeddine Z, Ghssein G. Overview of Yersinia pestis Metallophores: Yersiniabactin and Yersinopine. Biology. 2023; 12(4):598. https://doi.org/10.3390/biology12040598
Chicago/Turabian StyleChaaban, Taghrid, Yehya Mohsen, Zeinab Ezzeddine, and Ghassan Ghssein. 2023. "Overview of Yersinia pestis Metallophores: Yersiniabactin and Yersinopine" Biology 12, no. 4: 598. https://doi.org/10.3390/biology12040598





