Ectopic Expression of a Wheat R2R3-Type MYB Gene in Transgenic Tobacco Enhances Osmotic Stress Tolerance via Maintaining ROS Balance and Improving Root System Architecture
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Osmotic Stress Treatments
2.2. Isolation of TaMYB20 Gene
2.3. Semi-Quantitative RT-PCR
2.4. Plant Transformation
2.5. Stress Assay Analysis of the Transgenic Tobacco Plants
2.6. RSA Trait Measurements for WT Plants and Transgenic Tobacco Lines
2.7. Histochemical In Situ Detection of ROS, ∆Ψm, O•− and H2O2
2.8. Measurement of RWC and Ion Leakage
2.9. Assays of Antioxidant Enzymes Activity
2.10. HPLC Analysis of the Phenolic Compounds of Wild and Transgenic Plants
2.11. Statistical Analysis
3. Results
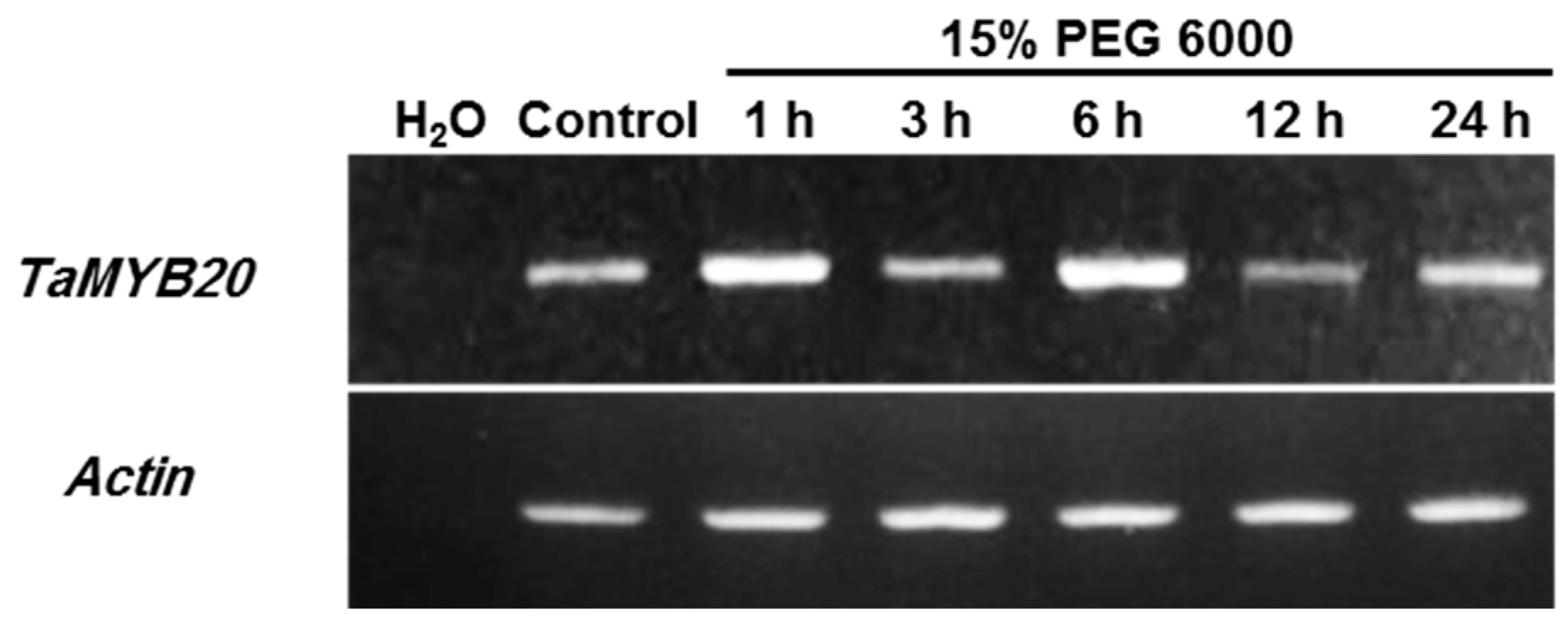
3.1. Expression Profiles of TaMYB20 in Wheat
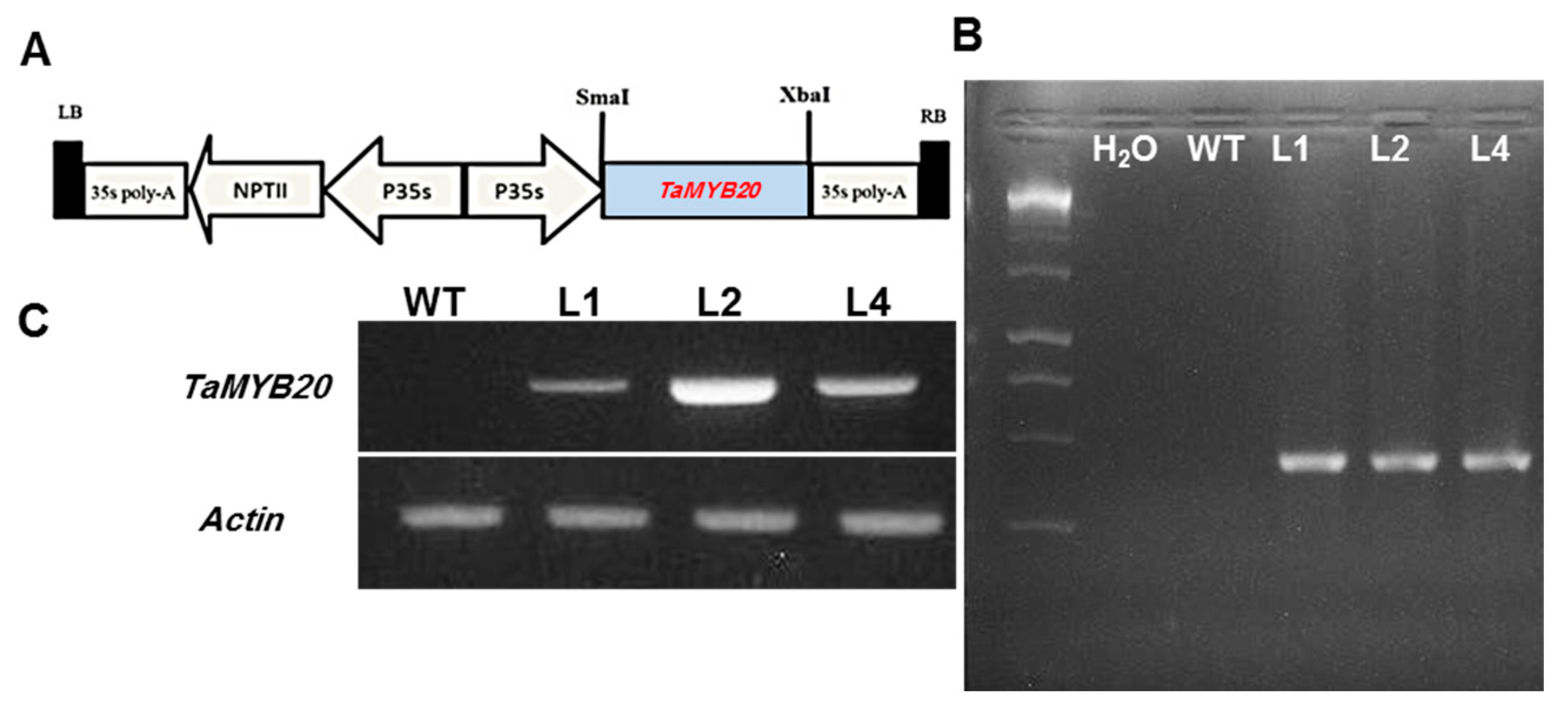
3.2. Isolation of TaMYB20 Gene and Generation of Transgenic Tobacco Plants
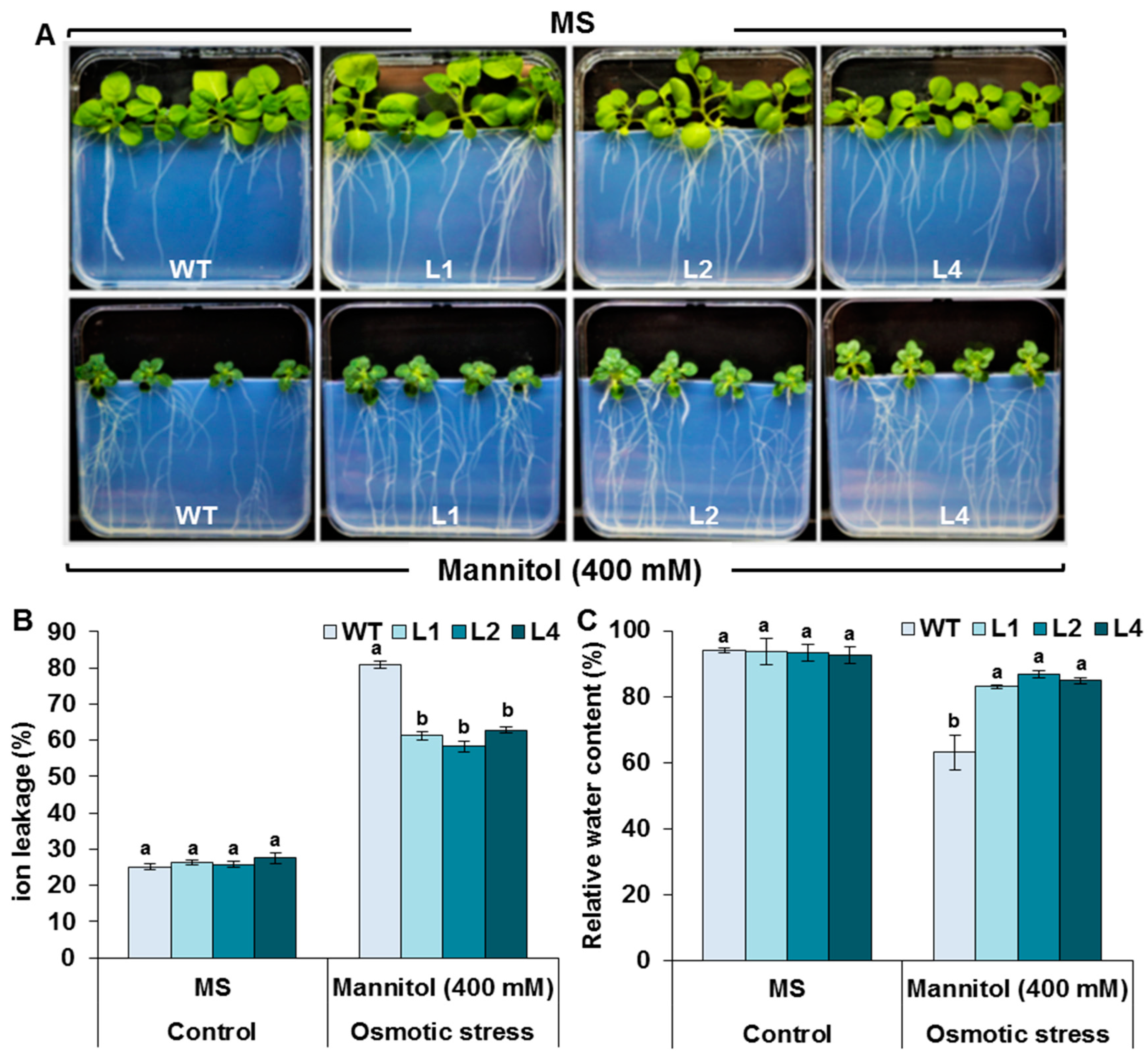
3.3. Evaluation of Stress Tolerance under In Vitro Conditions
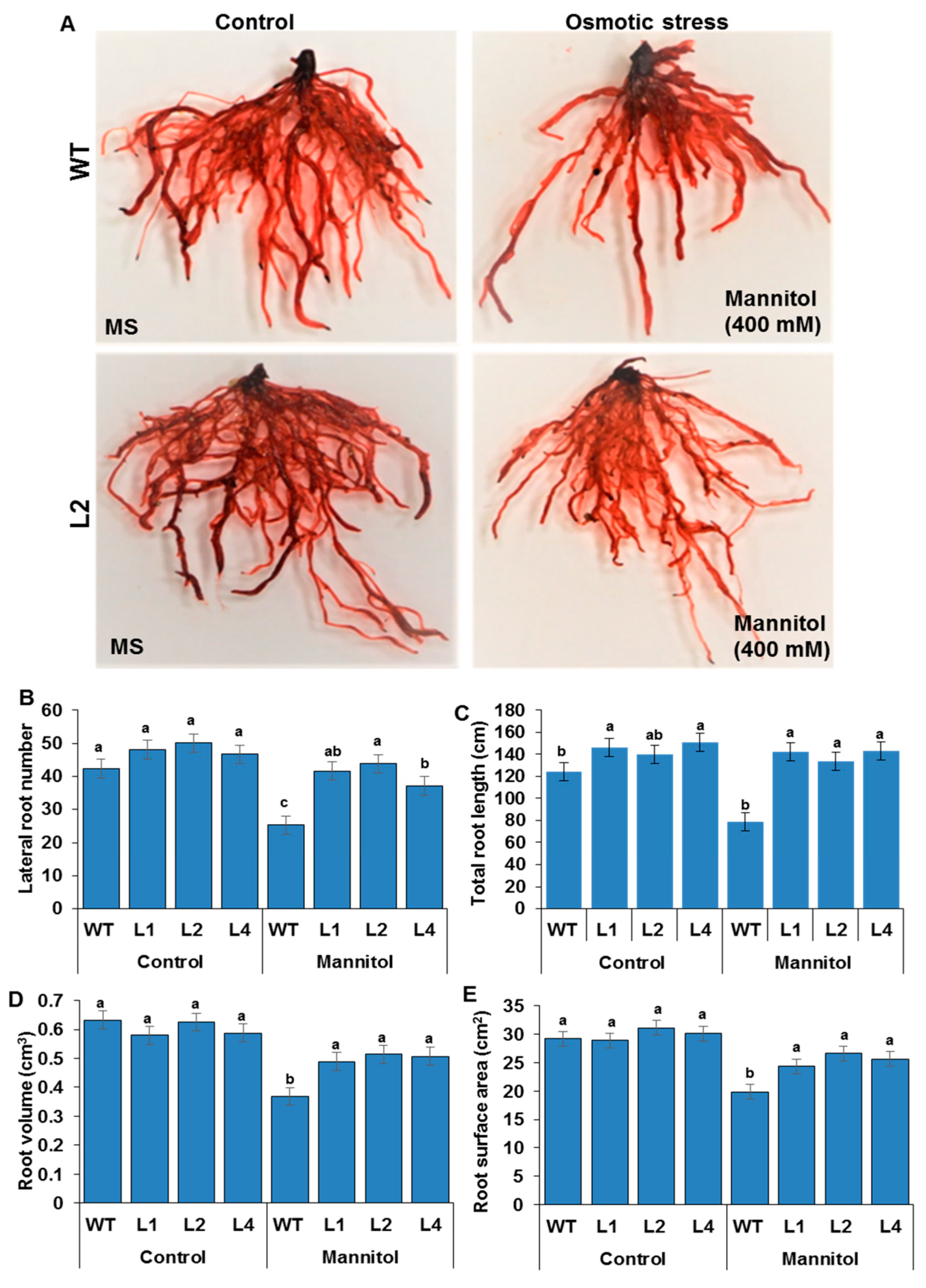
3.4. RSA Traits Measurements
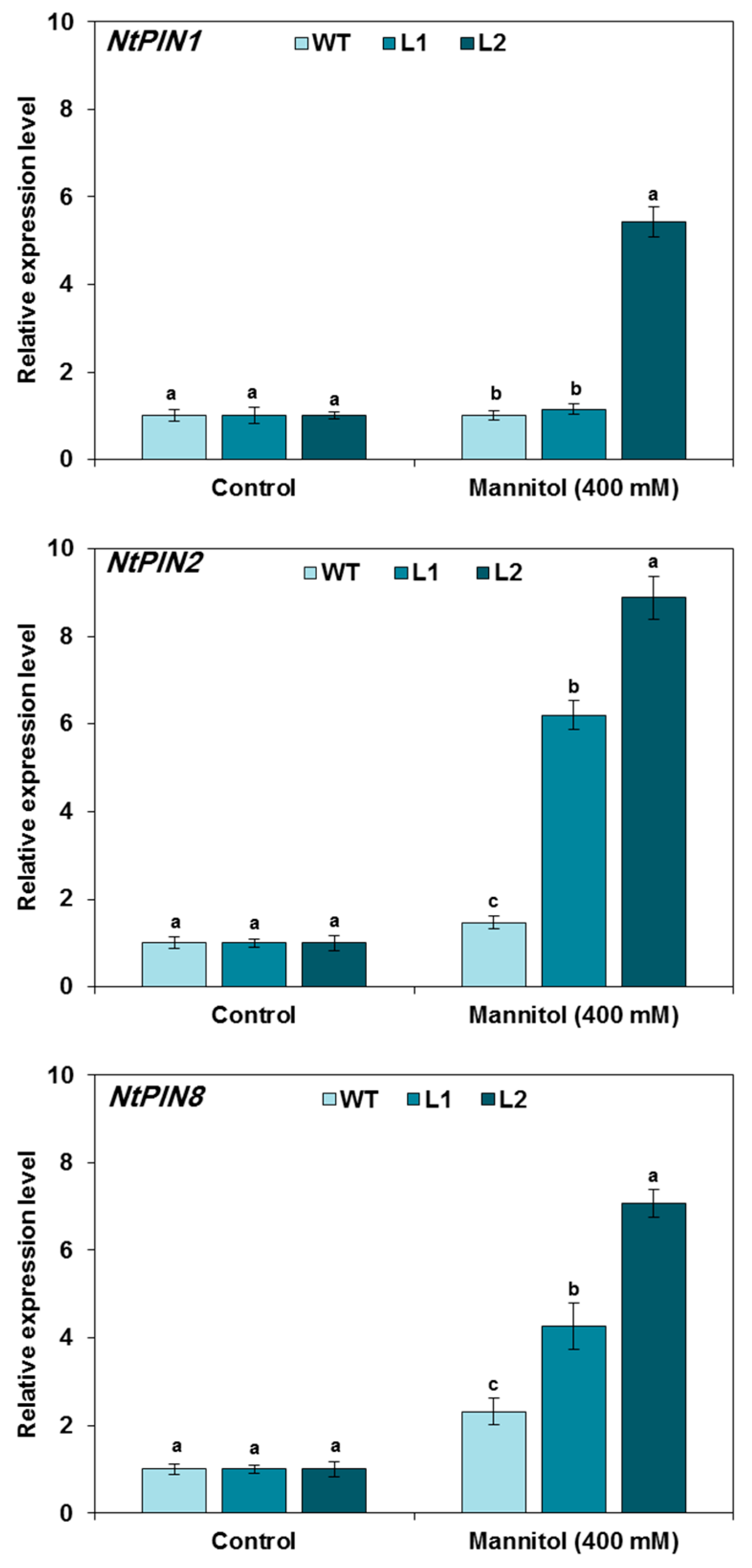
3.5. Expression of Auxin-Related Genes in TaMYB20 Tobacco Plants
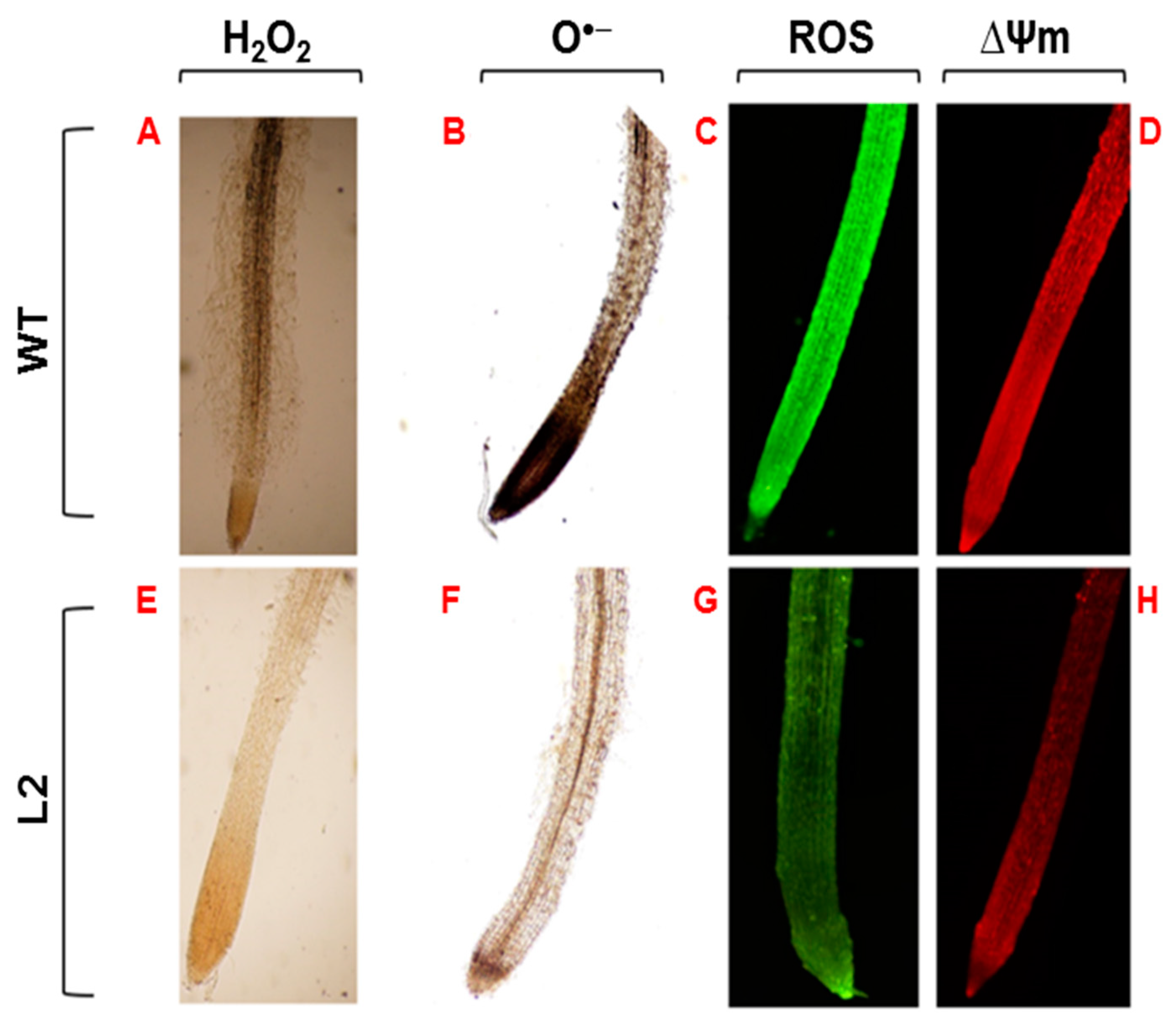
3.6. Histochemical In Situ Detection of ROS, ∆Ψm, O•− and H2O2
3.7. Effects of Drought Stress on RWC and IL
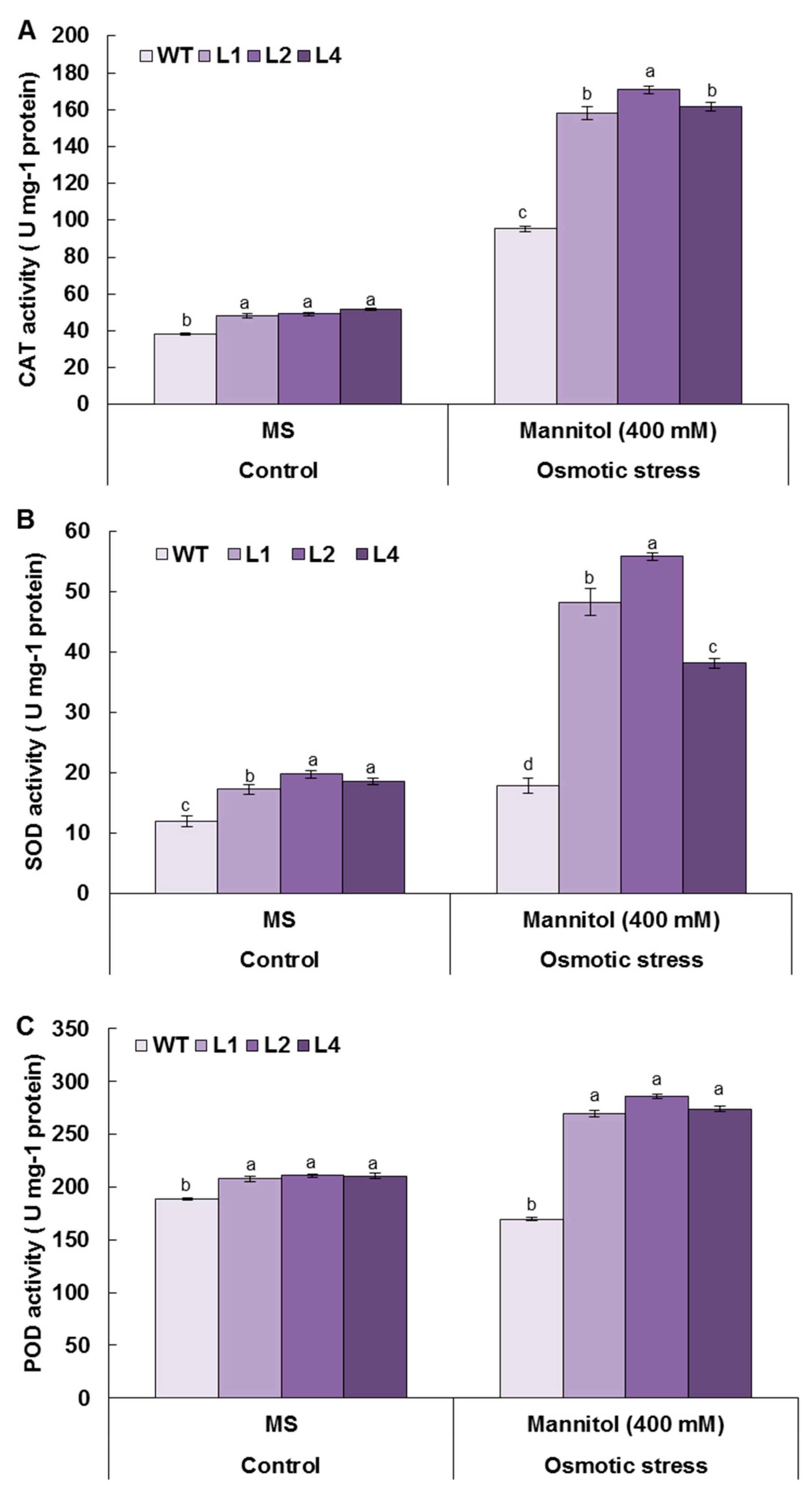
3.8. Overexpression of TaMYB20 Improves Antioxidant Enzymes Activities under Osmotic Conditions
3.9. Phenolic Compounds of Wild and Transgenic Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Du, H.; Zhang, L.; Liu, L.; Tang, X.F.; Yang, W.J.; Wu, Y.M.; Huang, Y.B.; Tang, Y.X. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 2009, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yang, H.; Zhang, X.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005, 15, 1196–1200. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Z.; Kang, J.; Kang, D.; Gu, H.; Qin, G. AtMYB14 Regulates Cold Tolerance in Arabidopsis. Plant Mol. Biol. Report. 2013, 31, 87–97. [Google Scholar] [CrossRef]
- Chen, T.; Li, W.; Hu, X.; Guo, J.; Liu, A.; Zhang, B. A Cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought Stress. Plant Cell Physiol. 2015, 56, 917–929. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Shi, Q.; Ren, Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020, 291, 110356. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; He, S.; Zhai, H.; Zhao, N.; Xing, S.; Wei, Z.; Liu, Q. A novel Sweetpotato transcription factor gene IbMYB116 enhances drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 1025. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, N.; Yang, J.; Guo, H.; Liu, Z.; Zheng, X.; Li, S.; Xiang, F. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020, 291, 110326. [Google Scholar] [CrossRef]
- Lee, T.G.; Jang, C.S.; Kim, J.Y.; Kim, D.S.; Park, J.H.; Kim, D.Y.; Seo, Y.W. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: Roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Plant. 2007, 129, 375–385. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, G.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol. 2014, 55, 1802–1812. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, F.; Sun, F.; Luo, Q.; Wang, R.; Hu, R.; Chen, M.; Chang, J.; Yang, G.; He, G. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 2017, 265, 112–123. [Google Scholar] [CrossRef]
- Wei, Q.; Luo, Q.; Wang, R.; Zhang, F.; He, Y.; Zhang, Y.; Qiu, D.; Li, K.; Chang, J.; Yang, G.; et al. A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt Stress responses in transgenic tobacco plants. Front. Plant Sci. 2017, 8, 1374. [Google Scholar] [CrossRef]
- Janiak, A.; Kwasniewski, M.; Szarejko, I. Gene expression regulation in roots under drought. J. Exp. Bot. 2016, 67, 1003–1014. [Google Scholar] [CrossRef]
- Azab, O.; Al-Doss, A.; Alshahrani, T.; El-Hendawy, S.; Zakri, A.M.; Abd-ElGawad, A.M. Root system architecture plasticity of bread wheat in response to oxidative burst under extended osmotic stress. Plants 2021, 10, 939. [Google Scholar] [CrossRef]
- Grzesiak, M.T.; Hordynska, N.; Maksymowicz, A.; Grzesiak, S.; Szechynska-Hebda, M. Variation among spring wheat (Triticum aestivum L.) genotypes in response to the drought stress. II-root system structure. Plants 2019, 8, 584. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Horsch, R.B.; Rogers, S.G.; Fraley, R.T. Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 1985, 50, 433–437. [Google Scholar] [CrossRef]
- Saquib, Q.; Musarrat, J.; Siddiqui, M.A.; Dutta, S.; Dasgupta, S.; Giesy, J.P.; Al-Khedhairy, A.A. Cytotoxic and necrotic responses in human amniotic epithelial (WISH) cells exposed to organophosphate insecticide phorate. Mutat. Res. 2012, 744, 125–134. [Google Scholar] [CrossRef]
- González, L.; González-Vilar, M. Determination of Relative Water Content. In Handbook of Plant Ecophysiology Techniques; Reigosa Roger, M.J., Ed.; Springer: Dordrecht, The Netherlands, 2001; pp. 207–212. [Google Scholar]
- Ben Romdhane, W.; Ben Saad, R.; Meynard, D.; Verdeil, J.L.; Azaza, J.; Zouari, N.; Fki, L.; Guiderdoni, E.; Al-Doss, A.; Hassairi, A. Ectopic expression of Aeluropus littoralis plasma membrane protein gene AlTMP1 confers abiotic stress tolerance in transgenic tobacco by improving water status and cation homeostasis. Int. J. Mol. Sci. 2017, 18, 692. [Google Scholar] [CrossRef]
- Abd-ElGawad, A.M.; Elshamy, A.I.; Al-Rowaily, S.L.; El-Amier, Y.A. Habitat affects the chemical profile, allelopathy, and antioxidant properties of essential oils and phenolic enriched extracts of the invasive plant Heliotropium Curassavicum. Plants 2019, 8, 482. [Google Scholar] [CrossRef]
- Ma, Q.H.; Wang, C.; Zhu, H.H. TaMYB4 cloned from wheat regulates lignin biosynthesis through negatively controlling the transcripts of both cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase genes. Biochimie 2011, 93, 1179–1186. [Google Scholar] [CrossRef]
- Zhao, Y.; Cheng, X.; Liu, X.; Wu, H.; Bi, H.; Xu, H. The wheat MYB transcription factor TaMYB(31) is involved in drought stress responses in Arabidopsis. Front. Plant Sci. 2018, 9, 1426. [Google Scholar] [CrossRef]
- Blanco, E.; Curci, P.L.; Manconi, A.; Sarli, A.; Zuluaga, D.L.; Sonnante, G. R2R3-MYBs in durum wheat: Genome-wide Identification, Poaceae-specific clusters, expression, and regulatory dynamics under abiotic stresses. Front. Plant Sci. 2022, 13, 896945. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Voss, U.; Harding, S.A.; Fannon, J.; Moody, L.A.; Yamada, E.; Swarup, K.; Nibau, C.; Bassel, G.W.; Choudhary, A.; et al. AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol. 2014, 203, 1194–1207. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Marcos, M.; Desvoyes, B.; Manzano, C.; Liberman, L.M.; Benfey, P.N.; Del Pozo, J.C.; Gutierrez, C. Control of Arabidopsis lateral root primordium boundaries by MYB36. New Phytol. 2017, 213, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Lu, M.Q.; Wang, Y.F.; Wang, Y.R.; Liu, Z.J.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 2012, 63, 5873–5885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.X.; Yao, P.F.; Zhao, J.L.; Wu, H.L.; Wang, S.; Chen, Y.; Hu, M.F.; Wang, T.; Li, C.L.; Wu, Q. A novel R2R3-MYB transcription factor FtMYB22 negatively regulates salt and drought stress through ABA-dependent pathway. Int. J. Mol. Sci. 2022, 23, 14549. [Google Scholar] [CrossRef]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W.; et al. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic Rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.B.; Ren, X.Q.; Zhang, F.; Qi, M.Y.; Zhao, H.Y.; Chen, X.H.; Ye, Y.X.; Yang, J.Y.; Li, S.G.; Zhang, Y.; et al. A R2R3-type MYB transcription factor gene from soybean, GmMYB12, is involved in flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis. Plant Biotechnol. Rep. 2019, 13, 219–233. [Google Scholar] [CrossRef]
- Overvoorde, P.; Fukaki, H.; Beeckman, T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010, 2, a001537. [Google Scholar] [CrossRef]
- Armengot, L.; Marques-Bueno, M.M.; Jaillais, Y. Regulation of polar auxin transport by protein and lipid kinases. J. Exp. Bot. 2016, 67, 4015–4037. [Google Scholar] [CrossRef]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazimalova, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef]
- Zhang, Y.; He, P.; Yang, Z.; Huang, G.; Wang, L.; Pang, C.; Xiao, H.; Zhao, P.; Yu, J.; Xiao, G. A genome-scale analysis of the PIN gene family reveals its functions in Cotton fiber development. Front. Plant Sci. 2017, 8, 461. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Guo, X.; Xu, F.; Wu, D.; Zhang, X.; Lou, M.; Luo, F.; Xu, G.; Zhang, Y. Overexpression of OsPIN2 regulates root growth and formation in response to phosphate deficiency in rice. Int. J. Mol. Sci. 2019, 20, 5144. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ganguly, A.; Lee, R.D.; Park, M.; Cho, H.T. Intracellularly Localized PIN-FORMED8 promotes lateral root Emergence in Arabidopsis. Front. Plant Sci. 2019, 10, 1808. [Google Scholar] [CrossRef]

| Primers | Sequence 5′-3′ |
|---|---|
| TaMYB20-F | CAGCGAACAGCGGCTCCCAAAAT |
| TaMYB20-R | GCACGGTCGTCGGGGTTTCCATT |
| TaMYB20-F | CCCGGGATGGGGAGGCAGCCGTGCT |
| TaMYB20-R | GCTCTAGATCACGGCCATGCTTCTTG |
| qTaMYB20-F | AAATTCCCGGGTCAGAAGGG |
| qTaMYB20-R | CATGCTTCTTGGTCGAAGCC |
| Taactin-F | AGTGGAGGTTCTACCATGTTTCCT |
| Taactin-R | CACTGTATTTCCTTTCAGGTGGTG |
| TaMYB20-F | CATGTACCTCCTCGGCATGG |
| TaMYB20-R | GGTAGTAGTGGTCGAACGGG |
| NtActin-F | TCCAGGACAAGGAGGGTAT |
| NtActin-R | CATCAACAACAGGCAACCTAG |
| qNtPIN1c-F | GCCTCCATTGCTGTTGGTCTA |
| qNtPIN1c-R | CAACAAAATGTAGTAAACCAAAGTGATA |
| qNtPIN2-F | AATAGTTTTGGAGGGGATGTTTTC |
| qNtPIN2-R | CCCCTTGTCTTCTTGTTGGTTC |
| qNtPIN8-F | TGACTTGGATCATAACAGGTCTTTC |
| qNtPIN8-R | AGATTCTTTAGTTGCATTGAGCTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azab, O.; Ben Romdhane, W.; El-Hendawy, S.; Ghazy, A.; Zakri, A.M.; Abd-ElGawad, A.M.; Al-Doss, A. Ectopic Expression of a Wheat R2R3-Type MYB Gene in Transgenic Tobacco Enhances Osmotic Stress Tolerance via Maintaining ROS Balance and Improving Root System Architecture. Biology 2024, 13, 128. https://doi.org/10.3390/biology13020128
Azab O, Ben Romdhane W, El-Hendawy S, Ghazy A, Zakri AM, Abd-ElGawad AM, Al-Doss A. Ectopic Expression of a Wheat R2R3-Type MYB Gene in Transgenic Tobacco Enhances Osmotic Stress Tolerance via Maintaining ROS Balance and Improving Root System Architecture. Biology. 2024; 13(2):128. https://doi.org/10.3390/biology13020128
Chicago/Turabian StyleAzab, Omar, Walid Ben Romdhane, Salah El-Hendawy, Abdelhalim Ghazy, Adel M. Zakri, Ahmed M. Abd-ElGawad, and Abdullah Al-Doss. 2024. "Ectopic Expression of a Wheat R2R3-Type MYB Gene in Transgenic Tobacco Enhances Osmotic Stress Tolerance via Maintaining ROS Balance and Improving Root System Architecture" Biology 13, no. 2: 128. https://doi.org/10.3390/biology13020128







