General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity
Abstract
:Simple Summary
Abstract
1. Introduction
2. Bacteriology of K. pneumoniae
3. Pathophysiology of K. pneumoniae
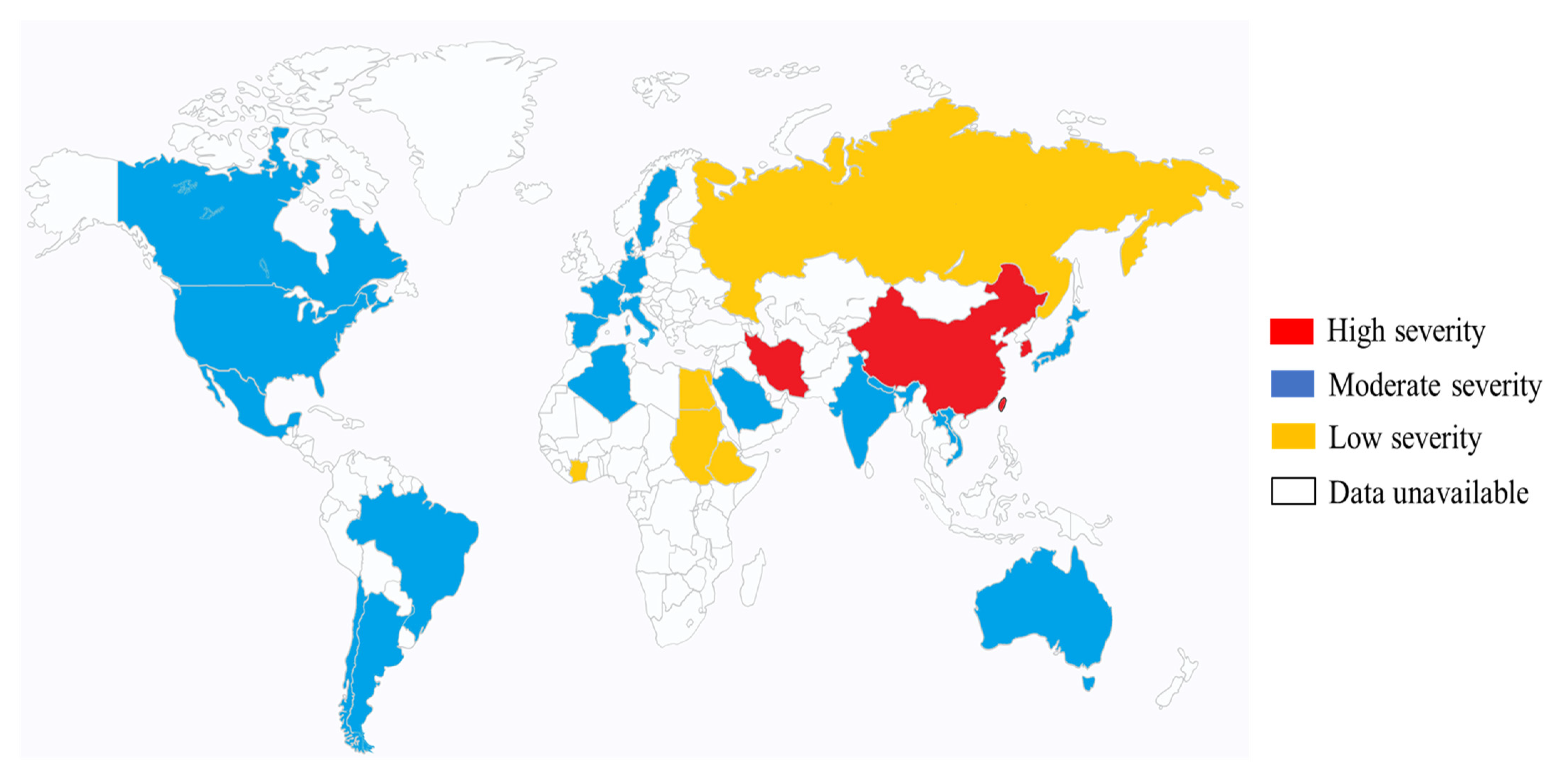
4. Epidemiology of K. pneumoniae (Mainly Hypervirulent K. pneumoniae in Eastern Asia)
5. Siderophores
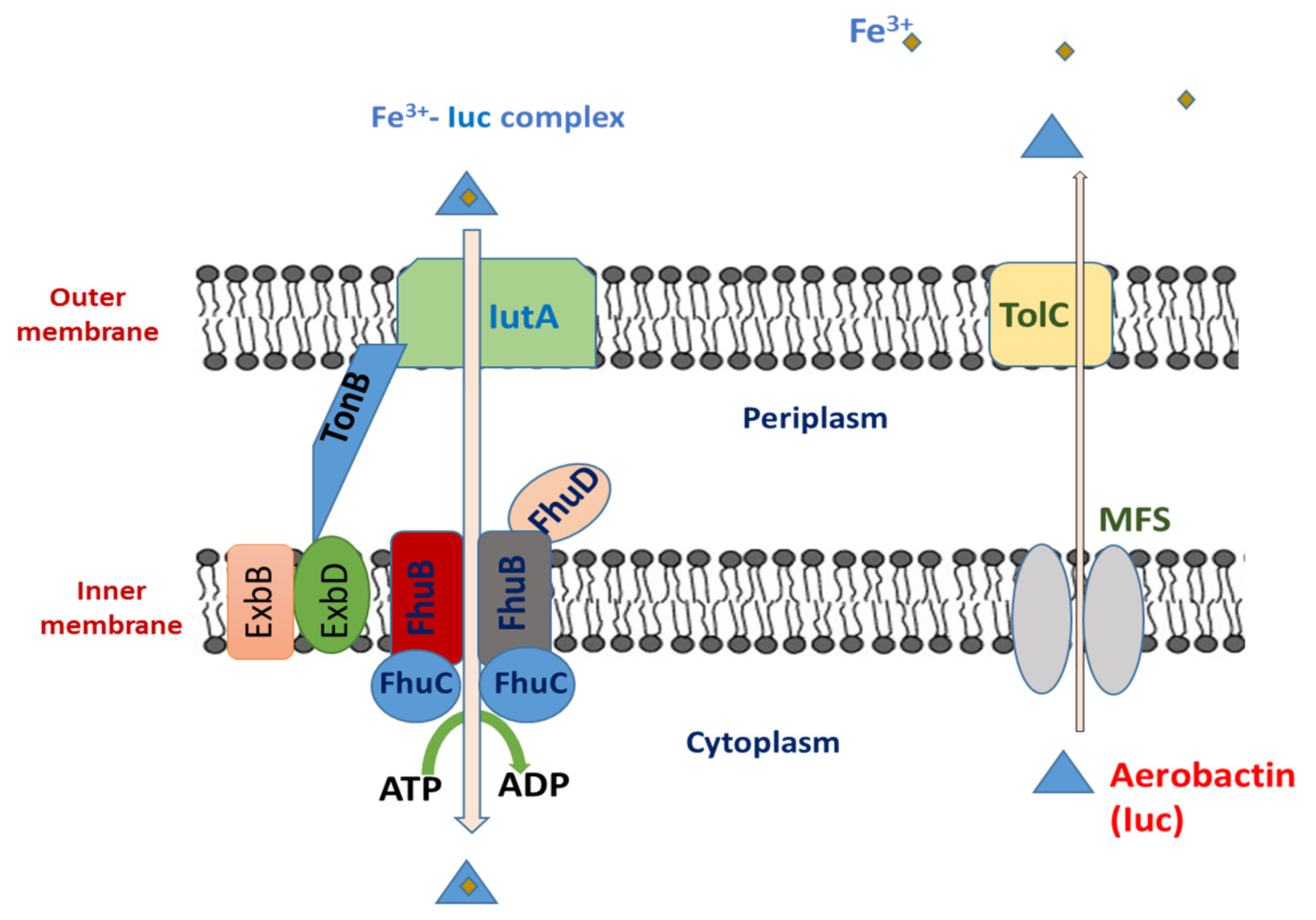
5.1. Aerobactin
5.1.1. Aerobactin Biosynthesis
5.1.2. Genetic Regulation
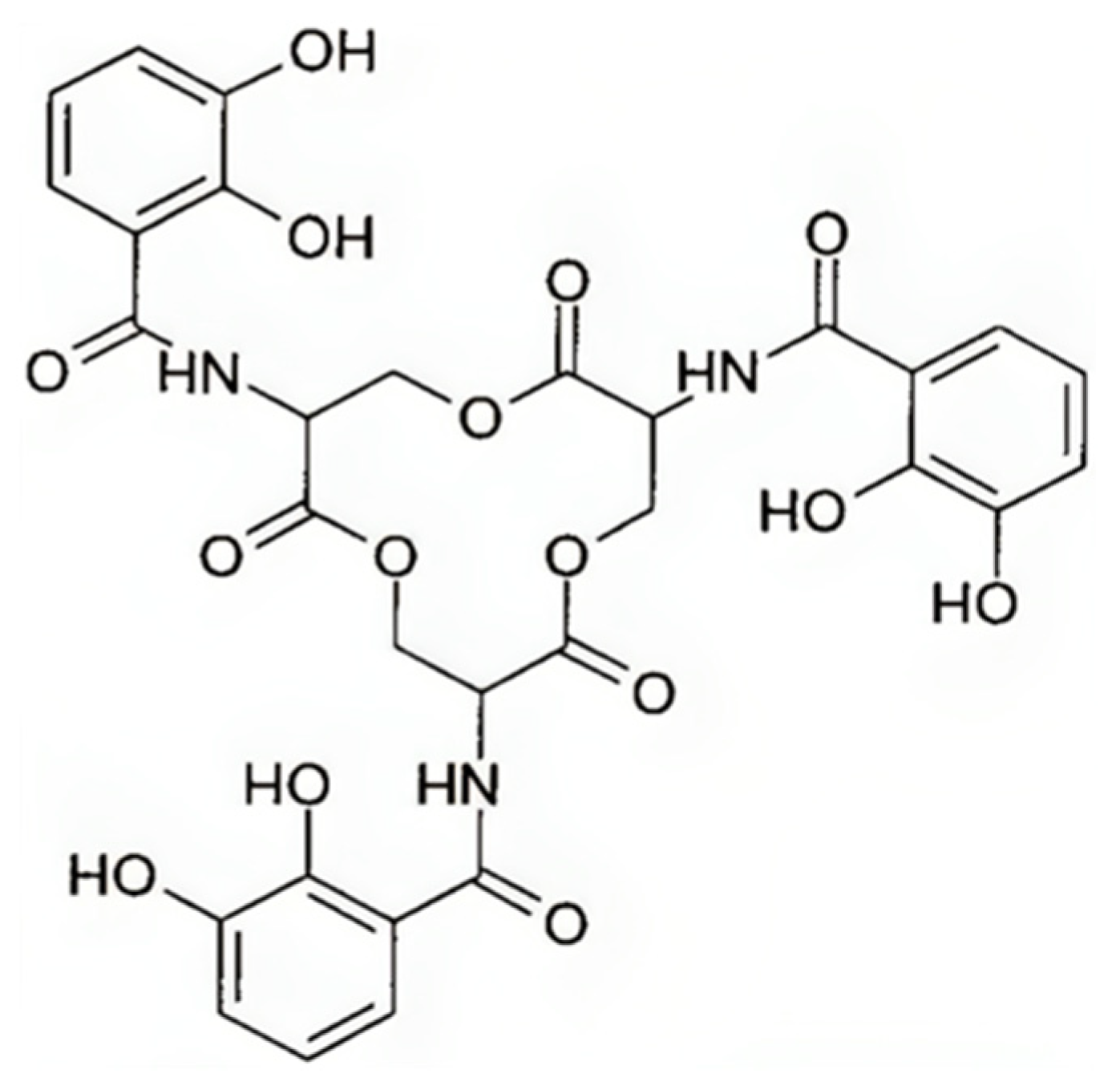
5.2. Enterobactin
5.2.1. Enterobactin Biosynthesis
5.2.2. Genetic Regulation
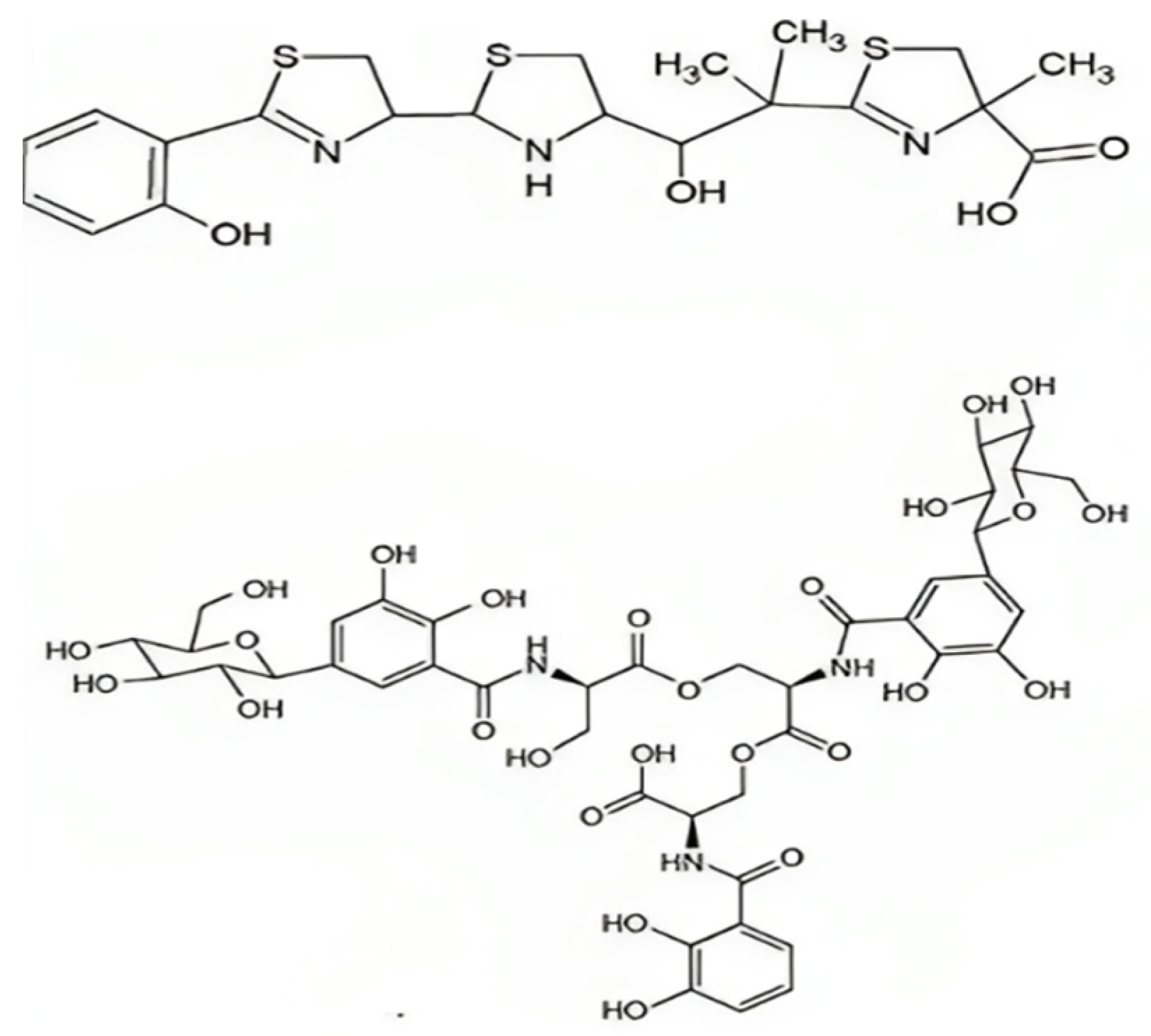
5.3. Salmochelin
5.4. Yersiniabactin
5.5. Siderophores and Virulence
5.6. Heme Uptake System hmuRSTUV
6. Mortality and Morbidity of K. pneumonia
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae-clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017.
- Magill, S.S.; O’Leary, E.; Janelle, S.J.; Thompson, D.L.; Dumyati, G.; Nadle, J.; Wilson, L.E.; Kainer, M.A.; Lynfield, R.; Greissman, S.; et al. Changes in Prevalence of Health Care-Associated Infections in U.S. Hospitals. N. Engl. J. Med. 2018, 379, 1732–1744. [Google Scholar] [CrossRef]
- Pomakova, D.K.; Hsiao, C.-B.; Beanan, J.M.; Olson, R.; MacDonald, U.; Keynan, Y.; Russo, T.A. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: An emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Shi, Q.; Zhang, P.; Chen, Y.; Yan, R.; Hua, X.; Jiang, Y.; Zhou, J.; Yu, Y. Core genome allelic profiles of clinical Klebsiella pneumoniae strains using a random forest algorithm based on multilocus sequence typing scheme for hypervirulence analysis. J. Infect. Dis. 2020, 221, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Shon, A.S.; Bajwa, R.P.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Chuang, Y.C.; Yu, W.L.; Lee, N.Y.; Chang, C.M.; Ko, N.Y.; Wang, L.R.; Ko, W.C. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: Association with invasive syndrome in patients with community-acquired bacteraemia. J. Intern. Med. 2006, 259, 606–614. [Google Scholar] [CrossRef]
- Li, W.; Sun, G.; Yu, Y.; Li, N.; Chen, M.; Jin, R.; Jiao, Y.; Wu, H. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 2014, 58, 225–232. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.Y.; Wan, L.G.; Jiang, W.Y.; Yang, J.H.; Li, F.Q. Virulence and transferability of resistance determinants in a novel Klebsiella pneumonia sequence type 1137 in China. Microb. Drug Resist. 2014, 20, 150–155. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, T.; Chen, L.; Du, H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 642484. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review. Biology 2022, 11, 1525. [Google Scholar] [CrossRef]
- Lamb, A.L. Breaking a pathogen’s iron will: Inhibiting siderophore production as an antimicrobial strategy. Biochim. Biophys. Acta 2015, 1854, 1054–1070. [Google Scholar] [CrossRef]
- Wand, M.E.; Baker, K.S.; Benthall, G.; McGregor, H.; McCowen, J.W.I.; DeheerGraham, A.; Sutton, J.M. Characterization of Pre-Antibiotic Era Klebsiella pneumoniae Isolates with Respect to Antibiotic/Disinfectant Susceptibility and Virulence in Galleria mellonella. Antimicrob. Agents Chemother. 2015, 59, 3966–3972. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Olson, R.; Macdonald, U.; Metzger, D.; Maltese, L.M.; Drake, E.J.; Gulick, A.M. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 2014, 82, 2356–2367. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Singh, P.; Srivastava, A. Synthesis, nature and utility of universal iron chelator-siderophore: A review. Microbiol. Res. 2018, 212–213, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Goetz, D.H.; Holmes, M.A.; Borregaard, N.; Bluhm, M.E.; Raymond, K.N.; Strong, R.K. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell. 2002, 10, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Lin, H.; Zhou, L.; Yu, Y.; Abergel, R.J.; Liu, D.R.; Raymond, K.N.; Wanner, B.L.; Strong, R.K.; Walsh, C.T.; et al. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. USA 2006, 103, 16502–16507. [Google Scholar] [CrossRef]
- Russo, T.A.; Olson, R.; Macdonald, U.; Beanan, J.; Davidson, B.A. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 2015, 83, 3325–3333. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Olson, R.; Fang, C.T.; Stoesser, N.; Miller, M.; MacDonald, U.; Hutson, A.; Barker, J.H.; La Hoz, R.M.; Johnson, J.R. Identification of biomarkers for the differentiation of hypervirulent Klebsiella pneumonia from classical K. pneumoniae. J. Clin. Microbiol. 2018, 56, e00776-18. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Wyres, K.L.; Judd, L.M.; Wick, R.R.; Jenney, A.; Brisse, S.; Holt, K.E. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018, 10, 77. [Google Scholar] [CrossRef]
- Bergey, D.H.; David, R.; Boone, G.M.; Garrity, R.W.C.; Bergey, D.H. Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA; London, UK, 2001. [Google Scholar]
- Wen, Z.; Zhang, J.-R. Chapter 3—Bacterial Capsules. In Molecular Medical Microbiology, 2nd ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Domenico, P.; Salo, R.J.; Cross, A.S.; Cunha, B.A. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumonia. Infect. Immun. 1994, 62, 4495–4499. [Google Scholar] [CrossRef]
- Brissac, T.; Martínez, E.; Kruckow, K.L.; Riegler, A.N.; Ganaie, F.; Im, H.; Bakshi, S.; Arroyo-Diaz, N.M.; Spencer, B.L.; Saad, J.S.; et al. Capsule Promotes Intracellular Survival and Vascular Endothelial Cell Translocation during Invasive Pneumococcal Disease. mBio 2021, 12, e0251621. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, M.S.; Handley, S.A.; Miller, V.L. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect. Immun. 2006, 74, 5402–5407. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Lin, T.L.; Chen, C.T.; Chen, Y.Y.; Hsieh, P.F.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 2015, 5, 15573. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shen, D.; Wu, H.; Ma, Y. Resistance of hypervirulent Klebsiella pneumoniae to both intracellular and extracellular killing of neutrophils. PLoS ONE 2017, 12, e0173638. [Google Scholar] [CrossRef] [PubMed]
- Shankar-Sinha, S.; Valencia, G.A.; Janes, B.K.; Rosenberg, J.K.; Whitfield, C.; Bender, R.A.; Standiford, T.J.; Younger, J.G. The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine pneumonia. Infect. Immun. 2004, 72, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Jhugroo, P. Antimicrobial Resistance, Mechanisms and Its Clinical Significance. Dis. Mon. 2020, 66, 100971. [Google Scholar] [CrossRef] [PubMed]
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Schroll, C.; Barken, K.B.; Krogfelt, K.A.; Struve, C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010, 10, 179. [Google Scholar] [CrossRef]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical Epidemiology of the Global Expansion of Klebsiella pneumoniae Carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Hsu, C.R.; Lin, T.L.; Pan, Y.J.; Hsieh, P.F.; Wang, J.T. Isolation of a Bacteriophage specific for a new capsular type of Klebsiella pneumoniae and Characterization of its Polysaccharide Depolymerase. PLoS ONE 2013, 8, e70092. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.I.; Picão, R.C.; Vespero, E.C.; Pelisson, M.; Zuleta, L.F.G.; Almeida, L.G.P.; Gerber, A.L.; Vasconcelos, A.T.R.; Gales, A.C.; Nicolás, M.F. Pyrosequencing-Based Analysis Reveals a Novel Capsular Gene Cluster in a KPC-Producing Klebsiella pneumoniae Clinical Isolate Identified in Brazil. BMC Microbiol. 2012, 12, 173. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, Y.; Liu, C.; Chen, Z.; Zhou, D. Molecular Pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014, 9, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.L.; Yang, Y.L.; Liao, P.C.; Chou, J.C.; Tsai, K.C.; Yang, A.S.; Sheu, F.; Lin, T.L.; Hsieh, P.F.; Wang, J.T.; et al. Structure and Immunological Characterization of the Capsular Polysaccharide of a Pyrogenic Liver Abscess caused by Klebsiella pneumoniae: Activation of Macrophages through Toll-like Receptor 4. J. Biol. Chem. 2011, 286, 21041–21051. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ferrer, S.; Peñaloza, H.F.; Budnick, J.A.; Bain, W.G.; Nordstrom, H.R.; Lee, J.S.; Van Tyne, D. Finding Order in the Chaos: Outstanding Questions in Klebsiella pneumoniae Pathogenesis. Infect. Immun. 2021, 89, e00693-20. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Altarriba, M.; Izquierdo, L.; Nogueras, M.M.; Regue, M.; Tomas, J.M. Cloning and Sequencing of the Klebsiella pneumoniae O5 wb Gene Cluster and its Role in Pathogenesis. Infect. Immun. 2000, 68, 2435–2440. [Google Scholar] [CrossRef]
- Clements, A.; Tull, D.; Jenney, A.W.; Farn, J.L.; Kim, S.H.; Bishop, R.E.; McPhee, J.B.; Hancock, R.E.; Hartland, E.L.; Pearse, M.J.; et al. Secondary Acylation of Klebsiella pneumoniae Lipopolysaccharide Contributes to Sensitivity to Antibacterial Peptides. J. Biol. Chem. 2007, 282, 15569–15577. [Google Scholar] [CrossRef]
- March, C.; Cano, V.; Moranta, D.; Llobet, E.; Pérez-Gutiérrez, C.; Tomás, J.M.; Suárez, T.; Garmendia, J.; Bengoechea, J.A. Role of Bacterial Surface Structures on the Interaction of Klebsiella pneumoniae with Phagocytes. PLoS ONE 2013, 8, e56847. [Google Scholar] [CrossRef]
- Struve, C.; Bojer, M.; Krogfelt, K.A. Characterization of Klebsiella pneumoniae Type 1 Fimbriae by Detection of Phase Variation during Colonization and Infection and Impact on Virulence. Infect. Immun. 2008, 76, 4055–4065. [Google Scholar] [CrossRef]
- Stahlhut, S.G.; Struve, C.; Krogfelt, K.A. Klebsiella pneumoniae Type 3 Fimbriae Agglutinate Yeast in a Mannose-Resistant Manner. J. Med. Microbiol. 2012, 61 Pt 3, 317–322. [Google Scholar] [CrossRef]
- Struve, C.; Bojer, M.; Krogfelt, K.A. Identification of a Conserved Chromosomal Region Encoding Klebsiella pneumoniae Type 1 and Type 3 Fimbriae and Assessment of the Role of Fimbriae in Pathogenicity. Infect. Immun. 2009, 77, 5016–5024. [Google Scholar] [CrossRef]
- Wu, C.C.; Huang, Y.J.; Fung, C.P.; Peng, H.L. Regulation of the Klebsiella pneumoniae Kpc Fimbriae by the Site-Specific Recombinase KpcI. Microbiology 2010, 156 Pt 7, 1983–1992. [Google Scholar] [CrossRef]
- Guerra, M.E.S.; Destro, G.; Vieira, B.; Lima, A.S.; Ferraz, L.F.C.; Hakansson, A.P.; Darrieux, M.; Converso, T.R. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Front. Cell. Infect. Microbiol. 2022, 12, 877995. [Google Scholar] [CrossRef]
- Sebghati, T.A.; Korhonen, T.K.; Hornick, D.B.; Clegg, S. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect. Immun. 1998, 66, 2887–2894. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, P.; Livrelli, V.; Sirot, D.; Joly, B.; Darfeuille-Michaud, A. A New Fimbrial Antigen Harbored by CAZ-5/ SHV-4-Producing Klebsiella pneumoniae Strains Involved in Nosocomial Infections. Infect. Immun. 1996, 64, 2266–2273. [Google Scholar] [CrossRef] [PubMed]
- Regueiro, V.; Moranta, D.; Frank, C.G.; Larrarte, E.; Margareto, J.; March, C.; Garmendia, J.; Bengoechea, J.A. Klebsiella pneumoniae Subverts the Activation of Inflammatory Responses in a NOD1-Dependent Manner. Cell Microbiol. 2011, 13, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Llobet, E.; March, C.; Gimenez, P.; Bengoechea, J.A. Klebsiella pneumoniae OmpA Confers Resistance to Antimicrobial Peptides. Antimicrob. Agents Chemother. 2009, 53, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.K.; Fung, C.P.; Lin, J.C.; Chen, J.-H.; Chang, F.-Y.; Chen, T.-L.; Siu, L.K. Klebsiella pneumoniae Outer Membrane Porins OmpK35 and OmpK36 Play Roles in Both Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2011, 55, 1485–1493. [Google Scholar] [CrossRef] [PubMed]
- Padilla, E.; Llobet, E.; Domenech-Sanchez, A.; Martinez-Martinez, L.; Bengoechea, J.A.; Alberti, S. Klebsiella pneumoniae AcrAB Efflux Pump Contributes to Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Pollack, M.; Nieman, R.; Reinhardt, J.; Charache, P.; Jett, M.; Hardy, P., Jr. Factors influencing colonisation and antibiotic-resistance patterns of gram-negative bacteria in hospital patients. Lancet 1972, 300, 668–671. [Google Scholar] [CrossRef]
- Chiu, C.T.; Lin, D.Y.; Liaw, Y.F. Metastatic Septic Endophthalmitis in Pyogenic Liver Abscess. J. Clin. Gastroenterol. 1988, 10, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C. Klebsiella pneumoniae Liver Abscess Associated With Septic Endophthalmitis. Arch. Intern. Med. 1986, 146, 1913. [Google Scholar] [CrossRef] [PubMed]
- Victor, L.Y.; Hansen, D.S.; Ko, W.C.; Sagnimeni, A.; Klugman, K.P.; Von Gottberg, A.; Goossens, H.; Wagener, M.M.; Benedi, V.J.; International Klebsiella Study Group. Virulence Characteristics of Klebsiella and Clinical Manifestations of K. pneumoniae Bloodstream Infections. Emerg. Infect. Dis. 2007, 13, 986–993. [Google Scholar]
- Coutinho, R.L.; Visconde, M.F.; Descio, F.J.; Nicoletti, A.G.; Pinto, F.C.; da Silva, A.C.R.; Rodrigues-Costa, F.; Gales, A.C.; Furtado, G.H. Community-acquired invasive liver abscess syndrome caused by a K1 serotype Klebsiella pneumoniae isolate in Brazil: A case report of hypervirulent ST23. Mem. Inst. Oswaldo Cruz 2014, 109, 970–971. [Google Scholar] [CrossRef] [PubMed]
- Fazili, T.; Sharngoe, C.; Endy, T.; Kiska, D.; Javaid, W.; Polhemus, M. Klebsiella pneumoniae Liver Abscess: An Emerging Disease. Am. J. Med. Sci. 2016, 351, 297–304. [Google Scholar] [CrossRef]
- Sturm, E.; Tai, A.; Lin, B.; Kwong, J.; Athan, E.; Howden, B.P.; Angliss, R.D.; Asaid, R.; Pollard, J. Bilateral osteomyelitis and liver abscess caused by hypervirulent Klebsiella pneumoniae—A rare clinical manifestation (case report). BMC Infect. Dis. 2018, 18, 1–5. [Google Scholar] [CrossRef]
- Sánchez-López, J.; García-Caballero, A.; Navarro-San Francisco, C.; Quereda, C.; Ruiz-Garbajosa, P.; Navas, E.; Dronda, F.; Morosini, M.I.; Cantón, R.; Diez-Aguilar, M. Hypermucoviscous Klebsiella pneumoniae: A challenge in community acquired infection. ID Cases 2019, 17, e00547. [Google Scholar] [CrossRef]
- Chung, D.R.; Lee, S.S.; Lee, H.R.; Kim, H.B.; Choi, H.J.; Eom, J.S.; Kim, J.S.; Choi, Y.H.; Lee, J.S.; Chung, M.H.; et al. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 2007, 54, 578–583. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, S.; Zhan, L.; Jin, Y.; Duan, J.; Hao, Z.; Lv, J.; Qi, X.; Chen, L.; Kreiswirth, B.N.; et al. Microbiological and Clinical Characteristics of Hypermucoviscous Klebsiella pneumoniae Isolates Associated with Invasive Infections in China. Front. Cell Infect. Microbiol. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Cubero, M.; Grau, I.; Tubau, F.; Pallarés, R.; Dominguez, M.; Liñares, J.; Ardanuy, C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin. Microbiol. Infect. 2016, 22, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Pitout, J.D.; Laupland, K.B.; Meatherall, B.; Gregson, D.B. Population-Based Surveillance for Hypermucoviscosity Klebsiella pneumoniae Causing Community-Acquired Bacteremia in Calgary, Alberta. Can. J. Infect. Dis. Med. Microbiol. 2013, 24, 61–64. [Google Scholar] [CrossRef]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumonia. Clin. Microbiol. Rev. 2019, 32, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Albasha, A.M.; Osman, E.H.; Abd-Alhalim, S.; Alshaib, E.F.; Al-Hassan, L.; Altayb, H.N. Detection of several carbapenems resistant and virulence genes in classical and hyper-virulent strains of Klebsiella pneumoniae isolated from hospitalized neonates and adults in Khartoum. BMC Res. Notes 2020, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Vasilyev, I.Y.; Nikolaeva, I.V.; Siniagina, M.N.; Kharchenko, A.M.; Shaikhieva, G.S. Multidrug-Resistant Hypervirulent Klebsiella pneumoniae Found Persisting Silently in Infant Gut Microbiota. Int. J. Microbiol. 2020, 2020, 4054393. [Google Scholar] [CrossRef]
- Sewunet, T.; Asrat, D.; Woldeamanuel, Y.; Ny, S.; Westerlund, F.; Aseffa, A.; Giske, C.G. High prevalence of blaCTX-M-15 and nosocomial transmission of hypervirulent epidemic clones of Klebsiella pneumoniae at a tertiary hospital in Ethiopia. JAC Antimicrob. Resist. 2021, 3, dlab001. [Google Scholar] [CrossRef]
- Emam, S.M.; Abdelrahman, S.; Hasan, A.A.; EL-Melouk, M.S. Hypervirulent Klebsiella pneumoniae at Benha University Hospitals. Egypt. J. Hosp. Med. 2023, 90, 3592–3597. [Google Scholar] [CrossRef]
- Liu, B.T.; Zhang, X.Y.; Wan, W.S.; Hao, J.J.; De Jiang, R.; Song, F.J. Characteristics of carbapenem-resistant Enterobacteriaceae in ready-to-eat vegetables in China. Front. Microbiol. 2018, 9, 359969. [Google Scholar] [CrossRef] [PubMed]
- Furlan, J.P.R.; Savazzi, E.A.; Stehling, E.G. Genomic insights into multidrug-resistant and hypervirulent Klebsiella pneumoniae co-harboring metal resistance genes in aquatic environments. Ecotoxicol. Environ. Saf. 2020, 201, 110782. [Google Scholar] [CrossRef]
- Gorrie, C.L.; Mirčeta, M.; Wick, R.R.; Edwards, D.J.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F.; Garlick, J.S.; Watson, K.M.; Pilcher, D.V.; et al. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin. Infect. Dis. 2017, 65, 208–215. [Google Scholar] [CrossRef]
- Fung, C.P.; Lin, Y.T.; Lin, J.C.; Chen, T.L.; Yeh, K.M.; Chang, F.Y.; Chuang, H.C.; Wu, H.S.; Tseng, C.P.; Siu, L.K. Klebsiella pneumoniae in Gastrointestinal Tract and Pyogenic Liver Abscess. Emerg. Infect. Dis. 2012, 18, 1322. [Google Scholar] [CrossRef]
- Harada, S.; Tateda, K.; Mitsui, H.; Hattori, Y.; Okubo, M.; Kimura, S.; Sekigawa, K.; Kobayashi, K.; Hashimoto, N.; Itoyama, S.; et al. Familial Spread of a Virulent Clone of Klebsiella pneumoniae Causing Primary Liver Abscess. J. Clin. Microbiol. 2011, 49, 2354–2356. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Hendrickson, E.L.; Tang, X.; Lux, R.; He, X.; Edlund, A.; McLean, J.S.; Shi, W. Klebsiella and Providencia Emerge as Lone Survivors Following Long-Term Starvation of Oral Microbiota. Proc. Natl. Acad. Sci. USA 2019, 116, 8499–8504. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic Analysis of Diversity, Population Structure, Virulence, and Antimicrobial Resistance in Klebsiella pneumoniae, an Urgent Threat to Public Health. Proc. Natl. Acad. Sci. USA 2015, 112, 3574–3581. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.F.; Lu, Y.R.; Lin, T.L.; Lai, L.Y.; Wang, J.T. Klebsiella pneumoniae Type VI Secretion System Contributes to Bacterial Competition, Cell Invasion, Type-1 Fimbriae Expression, and in vivo Colonization. J. Infect. Dis. 2019, 219, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Dunn, S.J.; Connor, C.; McNally, A. The Evolution and Transmission of Multi-Drug Resistant Escherichia coli and Klebsiella pneumoniae: The Complexity of Clones and Plasmids. Curr. Opin. Microbiol. 2019, 51, 51–56. [Google Scholar] [CrossRef]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchêne, S.; Jenney, A.; Holt, K.E. Distinct Evolutionary Dynamics of Horizontal Gene Transfer in Drug Resistant and Virulent Clones of Klebsiella pneumoniae. PLoS Genet. 2019, 15, 1008114. [Google Scholar] [CrossRef]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic Resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Monnet, D.L. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Stojowska-Swędrzyńska, K.; Łupkowska, A.; Kuczyńska-Wiśnik, D.; Laskowska, E. Antibiotic Heteroresistance in Klebsiella pneumoniae. Int. J. Mol. Sci. 2021, 23, 449. [Google Scholar] [CrossRef]
- Prince, S.E.; Dominger, K.A.; Cunha, B.A.; Klein, N.C. Klebsiella pneumoniae Pneumonia. Heart Lung 1997, 26, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Pana, Z.D.; Zaoutis, T. Treatment of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae (ESBLs) Infections: What Have We Learned Until Now? F1000Research 2018, 7. F1000 Faculty Rev-1347. [Google Scholar] [CrossRef] [PubMed]
- Fritzenwanker, M.; Imirzalioglu, C.; Herold, S.; Wagenlehner, F.M.; Zimmer, K.-P.; Chakraborty, T. Treatment Options for Carbapenem-Resistant Gram-Negative Infections. Dtsch. Arztebl. Int. 2018, 115, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, M.S.; O’Connor, C.; Miller, V.L. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 2007, 75, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.F.; Lin, T.L.; Lee, C.Z.; Tsai, S.F.; Wang, J.T. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2008, 197, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.C.; Drake, E.J.; Grant, T.D.; Gulick, A.M. Structural and Functional Characterization of Aerobactin Synthetase IucA from a Hypervirulent Pathotype of Klebsiella pneumoniae. Biochemistry 2016, 55, 3559–3570. [Google Scholar] [CrossRef] [PubMed]
- Mydy, L.S.; Bailey, D.C.; Patel, K.D.; Rice, M.R.; Gulick, A.M. The Siderophore Synthetase IucA of the Aerobactin Biosynthetic Pathway Uses an Ordered Mechanism. Biochemistry 2020, 59, 2143–2153. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, M.; Stork, M. Plasmid-Encoded Iron Uptake Systems. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Gulick, A.M. Aerobactin Synthesis Proteins as Antivirulence Targets in Hypervirulent Klebsiella pneumoniae. ACS Infect. Dis. 2019, 5, 1052–1054. [Google Scholar] [CrossRef]
- Carroll, C.S.; Moore, M.M. Ironing out siderophore biosynthesis: A review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 356–381. [Google Scholar] [CrossRef]
- Braun, V. Energy-coupled transport and signal transduction through the gramnegative outer membrane via the TonB-ExbB-ExbD–dependent receptor proteins. FEMS Microbiol. Lett. 1995, 16, 295–307. [Google Scholar] [CrossRef]
- Bagg, A.; and Neilands, J.B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 1987, 51, 509–518. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, V.; Wee, S.; Herrero, M.; Neilands, J.B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 1987, 169, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Santander, J.; Golden, G.; Wanda, S.Y.; Curtiss, R. Fur-regulated iron uptake system of Edwardsiella ictaluri and its influence on pathogenesis and immunogenicity in the catfish host. Infect. Immun. 2012, 80, 2689–2703. [Google Scholar] [CrossRef]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.M.; Mori, I.; Walsh, C.T. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 1998, 37, 2648–2659. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.; Liu, J.; Rusnak, F.; Sakaitani, M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem. Rev. 1990, 90, 1105–1129. [Google Scholar] [CrossRef]
- Lambalot, R.H.; Gehring, A.M.; Flugel, R.S.; Zuber, P.; LaCelle, M.; Marahiel, M.A.; Reid, R.; Khosla, C.; Walsh, C.T. A new enzyme superfamily—The phosphopantetheinyl transferases. Chem. Biol. 1996, 3, 923–936. [Google Scholar] [CrossRef]
- Furrer, J.L.; Sanders, D.N.; Hook-Barnard, I.G.; McIntosh, M.A. Export of the siderophore enterobactin in Escherichia coli: Involvement of a 43 kDa membrane exporter. Mol. Microbiol. 2002, 44, 1225–1234. [Google Scholar] [CrossRef]
- Crosa, J.H.; Mey, A.R.; Payne, S.M. Iron Transport in Bacteria; ASM Press: Washington, DC, USA, 2004. [Google Scholar]
- Annamalai, R.; Jin, B.; Cao, Z.; Newton, S.M.; Klebba, P.E. Recognition of ferric catecholates by FepA. J. Bacteriol. 2004, 186, 3578–3589. [Google Scholar] [CrossRef]
- Buchanan, S.K.; Smith, B.S.; Venkatramani, L.; Xia, D.; Esser, L.; Palnitkar, M.; Chakraborty, R.; Van Der Helm, D.; Deisenhofer, J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 1999, 6, 56–63. [Google Scholar]
- Faraldo-Gomez, J.D.; Sansom, M.S. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 2003, 4, 105–116. [Google Scholar] [CrossRef]
- Lin, H.; Fischbach, M.A.; Liu, D.R.; Walsh, C.T. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 2005, 127, 11075–11084. [Google Scholar] [CrossRef]
- Mohsen, Y.; Tarchichi, N.; Barakat, R.; Kawtharani, I.; Ghandour, R.; Ezzeddine, Z.; Ghssein, G. The Different Types of Metallophores Produced by Salmonella enterica: A Review. Microbiol. Res. 2023, 14, 1457–1469. [Google Scholar] [CrossRef]
- Müller, S.; Valdebenito, M.; Hantke, K. Salmochelin, the longoverlooked catecholate siderophore of Salmonella. Biometals 2009, 22, 691–695. [Google Scholar] [CrossRef]
- Hantke, K.; Nicholson, G.; Rabsch, W.; Winkelmann, G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 2003, 100, 3677–3682. [Google Scholar] [CrossRef]
- Zhu, M.; Valdebenito, M.; Winkelmann, G.; Hantke, K. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 2005, 151, 2363–2372. [Google Scholar] [CrossRef]
- Lim, D.; Kim, K.; Song, M.; Jeong, J.H.; Chang, J.H.; Kim, S.R.; Hong, C.; Im, S.S.; Park, S.H.; Lee, J.C.; et al. Transcriptional regulation of Salmochelin glucosyltransferase by Fur in Salmonella. Biochem. Biophys. Res. Commun. 2020, 529, 70–76. [Google Scholar] [CrossRef]
- Chambers, C.E.; McIntyre, D.D.; Mouck, M.; Sokol, P.A. Physical and structural characterization of yersiniophore, a siderophore produced by clinical isolates of Yersinia enterocolitica. Biometals 1996, 9, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Bearden, S.W.; Fetherston, J.D.; Perry, R.D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 1997, 65, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.M.; DeMoll, E.; Fetherston, J.D.; Mori, I.; Mayhew, G.F.; Blattner, F.R.; Walsh, C.T.; Perry, R.D. Iron acquisition in plague: Modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 1998, 5, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Bobrov, A.G.; Geoffroy, V.A.; Perry, R.D. Yersiniabactin production requires the thioesterase domain of HMWP2 and YbtD, a putative phosphopantetheinylate transferase. Infect. Immun. 2002, 70, 4204–4214. [Google Scholar] [CrossRef] [PubMed]
- Chaaban, T.; Mohsen, Y.; Ezzeddine, Z.; Ghssein, G. Overview of Yersinia pestis Metallophores: Yersiniabactin and Yersinopine. Biology 2023, 12, 598. [Google Scholar] [CrossRef] [PubMed]
- Bachman, M.A.; Lenio, S.; Schmidt, L.; Oyler, J.E.; Weiser, J.N. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 2012, 3, e00224-11. [Google Scholar] [CrossRef] [PubMed]
- Price, S.L.; Vadyvaloo, V.; DeMarco, J.K.; Brady, A.; Gray, P.A.; Kehl-Fie, T.E.; Garneau-Tsodikova, S.; Perry, R.D.; Lawrenz, M.B. Yersiniabactin contributes to overcoming zinc restriction during Yersinia pestisinfection of mammalian and insect hosts. Proc. Natl. Acad. Sci. USA 2021, 118, e2104073118. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.D.; Balbo, P.B.; Jones, H.A.; Fetherston, J.D.; DeMoll, E. Yersiniabactin from Yersinia pestis: Biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 1999, 145, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Podschun, R.; Sievers, D.; Fischer, A.; Ullmann, U. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of Klebsiella isolates causing human urinary tract infections. J. Infect. Dis. 1993, 168, 1415–1421. [Google Scholar] [CrossRef]
- Tarkkanen, A.M.; Allen, B.L.; Williams, P.H.; Kauppi, M.; Haahtela, K.; Siitonen, A.; Orskov, I.; Orskov, F.; Clegg, S.; Korhonen, T.K. Fimbriation, capsulation, and iron-scavenging systems of Klebsiella strains associated with human urinary tract infection. Infect. Immun. 1992, 60, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Koczura, R.; Kaznowski, A. Occurrence of the Yersinia highpathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog. 2003, 35, 197–202. [Google Scholar] [CrossRef]
- El Fertas-Aissani, R.; Messai, Y.; Alouache, S.; Bakour, R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumonia strains isolated from different clinical specimens. Pathol. Biol. 2013, 61, 209–216. [Google Scholar] [CrossRef]
- Lai, Y.C.; Peng, H.L.; Chang, H.Y. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 2001, 69, 7140–7145. [Google Scholar] [CrossRef]
- Cowland, J.B.; Borregaard, N. Molecular characterization and pattern of tissue expression of the gene for neutrophil gelatinase-associated lipocalin from humans. Genomics 1997, 45, 17–23. [Google Scholar] [CrossRef]
- Chan, Y.R.; Liu, J.S.; Pociask, D.A.; Zheng, M.; Mietzner, T.A.; Berger, T.; Mak, T.W.; Clifton, M.C.; Strong, R.K.; Ray, P.; et al. Lipocalin 2 is required for pulmonary host defense against Klebsiella infection. J. Immunol. 2009, 182, 4947–4956. [Google Scholar] [CrossRef]
- Nelson, A.L.; Barasch, J.M.; Bunte, R.M.; Weiser, J.N. Bacterial colonization of nasal mucosa induces expression of siderocalin, an ironsequestering component of innate immunity. Cell Microbiol. 2005, 7, 1404–1417. [Google Scholar] [CrossRef]
- Bachman, M.A.; Miller, V.L.; Weiser, J.N. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog. 2009, 5, e1000622. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, S.I.; Soter, N.A.; Center, D.M.; Austen, K.F. Cold urticaria. Recognition and characterization of a neutrophil chemotactic factor which appears in serum during experimental cold challenge. J. Clin. Investig. 1977, 60, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Bachman, M.A.; Oyler, J.E.; Burns, S.H.; Caza, M.; Lepine, F.; Dozois, C.M.; Weiser, J.N. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 2011, 79, 3309–3316. [Google Scholar] [CrossRef] [PubMed]
- Struve, C.; Roe, C.C.; Stegger, M.; Stahlhut, S.G.; Hansen, D.S.; Engelthaler, D.M.; Andersen, P.S.; Driebe, E.M.; Keim, P.; Krogfelt, K.A. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 2015, 6, e00630-15. [Google Scholar] [CrossRef]
- Nassif, X.; Sansonetti, P.J. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 1986, 54, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.L.; Chiang, M.K.; Liou, W.J.; Chen, Y.T.; Peng, H.L.; Chiou, C.S.; Liu, K.S.; Lu, M.C.; Tung, K.C.; Lai, Y.C. Correlation between Klebsiella pneumonia carrying pLVPK-derived loci and abscess formation. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Systemic iron homeostasis. Physiol. Rev. 2013, 93, 1721–1741. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Lu, Y.; Yan, R.; Fang, L.; Zhao, D.; Jiang, Y.; Yu, Y.; Du, X.; Zhou, J. Development of a Novel Typing Scheme Based on the Genetic Diversity of Heme/Hemin Uptake System Hmu in Klebsiella pneumoniae Species Complex. Microbiol. Spectr. 2023, 11, e01062-22. [Google Scholar] [CrossRef] [PubMed]
- Schwiesow, L.; Mettert, E.; Wei, Y.; Miller, H.K.; Herrera, N.G.; Balderas, D.; Kiley, P.J.; Auerbuch, V. Control of hmu heme uptake genes in Yersinia pseudotuberculosis in response to iron sources. Front. Cell Infect. Microbiol. 2018, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Huang, X.; Rao, H.; Yu, H.; Long, S.; Li, Y.; Zhang, J. Klebsiella pneumoniae bacteremia mortality: A systematic review and meta-analysis. Front. Cell Infect. Microbiol. 2023, 13, 1157010. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, J.; Huang, F.; Fang, J.; Cao, Y.; Zhang, K.; Zhou, H.; Cai, J.; Cui, W.; Chen, C.; et al. Clinical characteristics, risk factors and outcomes of Klebsiella pneumoniae pneumonia developing secondary Klebsiella pneumoniae bloodstream infection. BMC Pulm. Med. 2023, 23, 102. [Google Scholar] [CrossRef] [PubMed]
- Santos, W.M.D.; Secoli, S.R. Economic burden of inpatients infected with Klebsiella pneumoniae carbapenemase. Einstein 2019, 17, eGS4444. [Google Scholar] [CrossRef]
- Ezzeddine, Z.; Ghssein, G. Towards new antibiotics classes targeting bacterial metallophores. Microb. Pathog. 2023, 182, 106221. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, R.; Chakkour, M.; Zein El Dine, H.; Obaseki, E.F.; Obeid, S.T.; Jezzini, A.; Ghssein, G.; Ezzeddine, Z. General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity. Biology 2024, 13, 78. https://doi.org/10.3390/biology13020078
Abbas R, Chakkour M, Zein El Dine H, Obaseki EF, Obeid ST, Jezzini A, Ghssein G, Ezzeddine Z. General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity. Biology. 2024; 13(2):78. https://doi.org/10.3390/biology13020078
Chicago/Turabian StyleAbbas, Rim, Mohamed Chakkour, Hiba Zein El Dine, Eseiwi Folorunsho Obaseki, Soumaya T. Obeid, Aya Jezzini, Ghassan Ghssein, and Zeinab Ezzeddine. 2024. "General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity" Biology 13, no. 2: 78. https://doi.org/10.3390/biology13020078





