First Observations of Buzzards (Buteo) as Definitive Hosts of Sarcocystis Parasites Forming Cysts in the Brain Tissues of Rodents in Lithuania
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
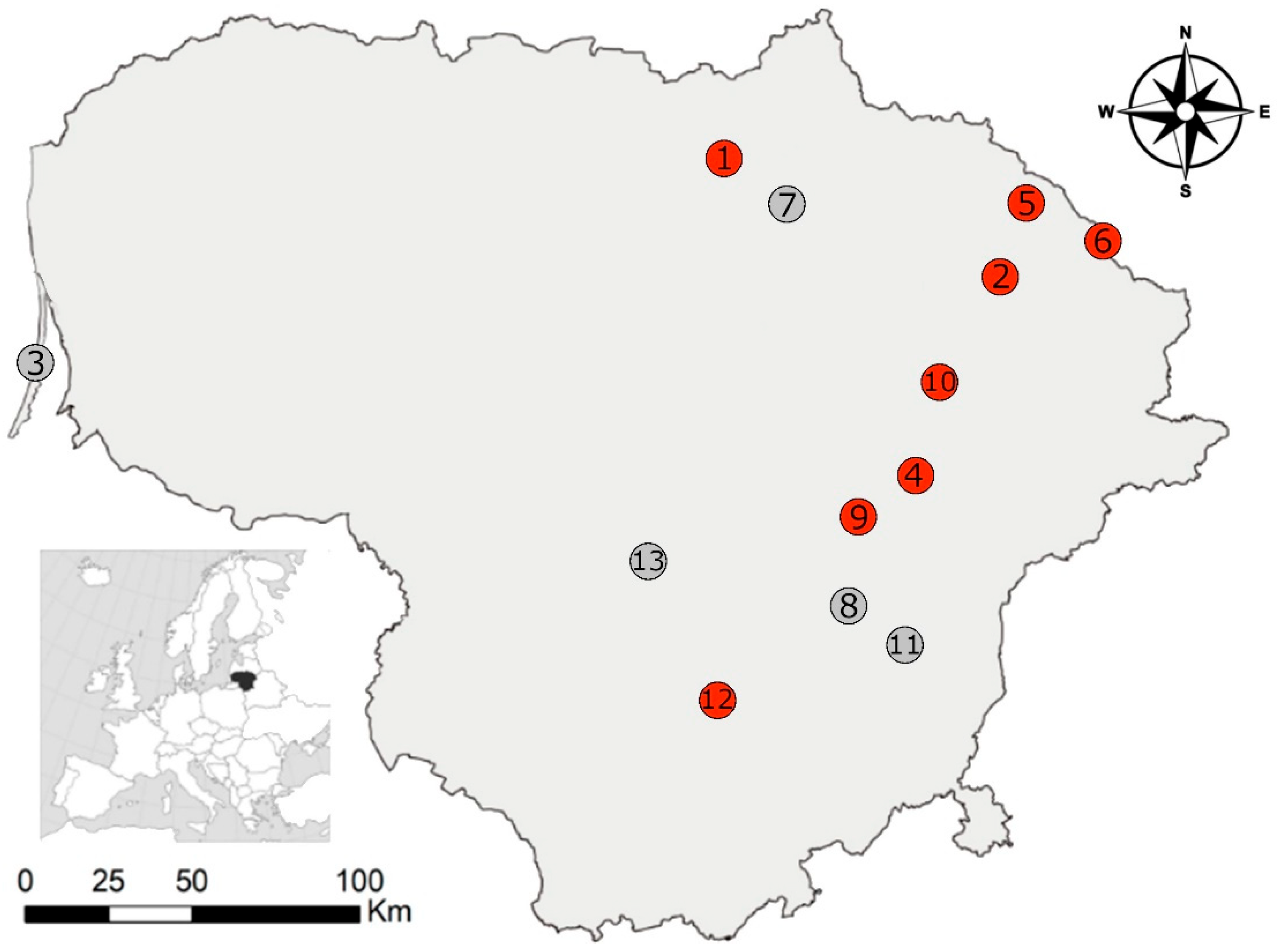
2.1. Trapping of Small Mammals
2.2. Morphological Examination of Sarcocystis spp. from Brain Tissues of Small Mammals
2.3. Genetic Characterization of Sarcocystis spp. Detected in Small Mammals
2.4. Collection of Buteo Buzzards
2.5. Morphological Examination of Sarcocystis spp. from Intestines of Buteo Buzzards
2.6. The Genetic Identification of Sarcocystis spp. from Intestines of Buteo Buzzards
2.7. Phylogenetic Analysis
2.8. Statistical Tests
3. Results
3.1. Prevalence and Parasite Load of Sarcocystis spp. in Small Mammals
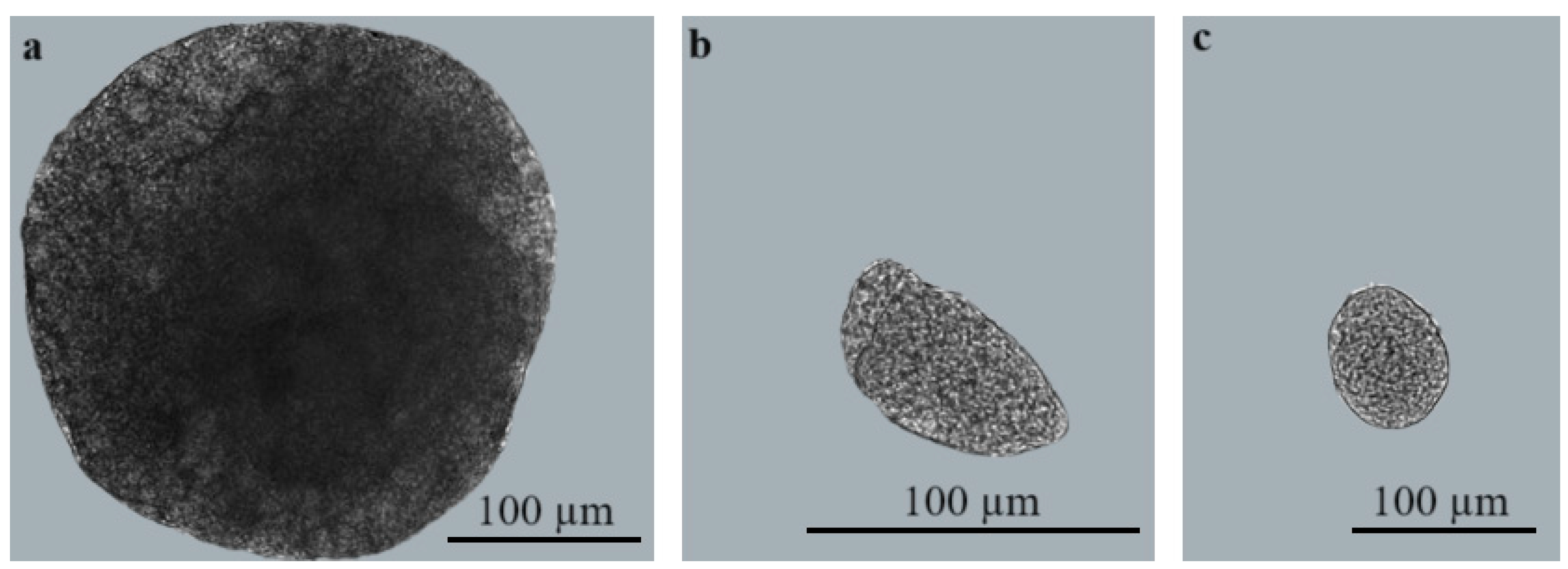
3.2. Morphological and Molecular Characterisation of Sarcocystis spp. in Brain Tissues of Small Mammals
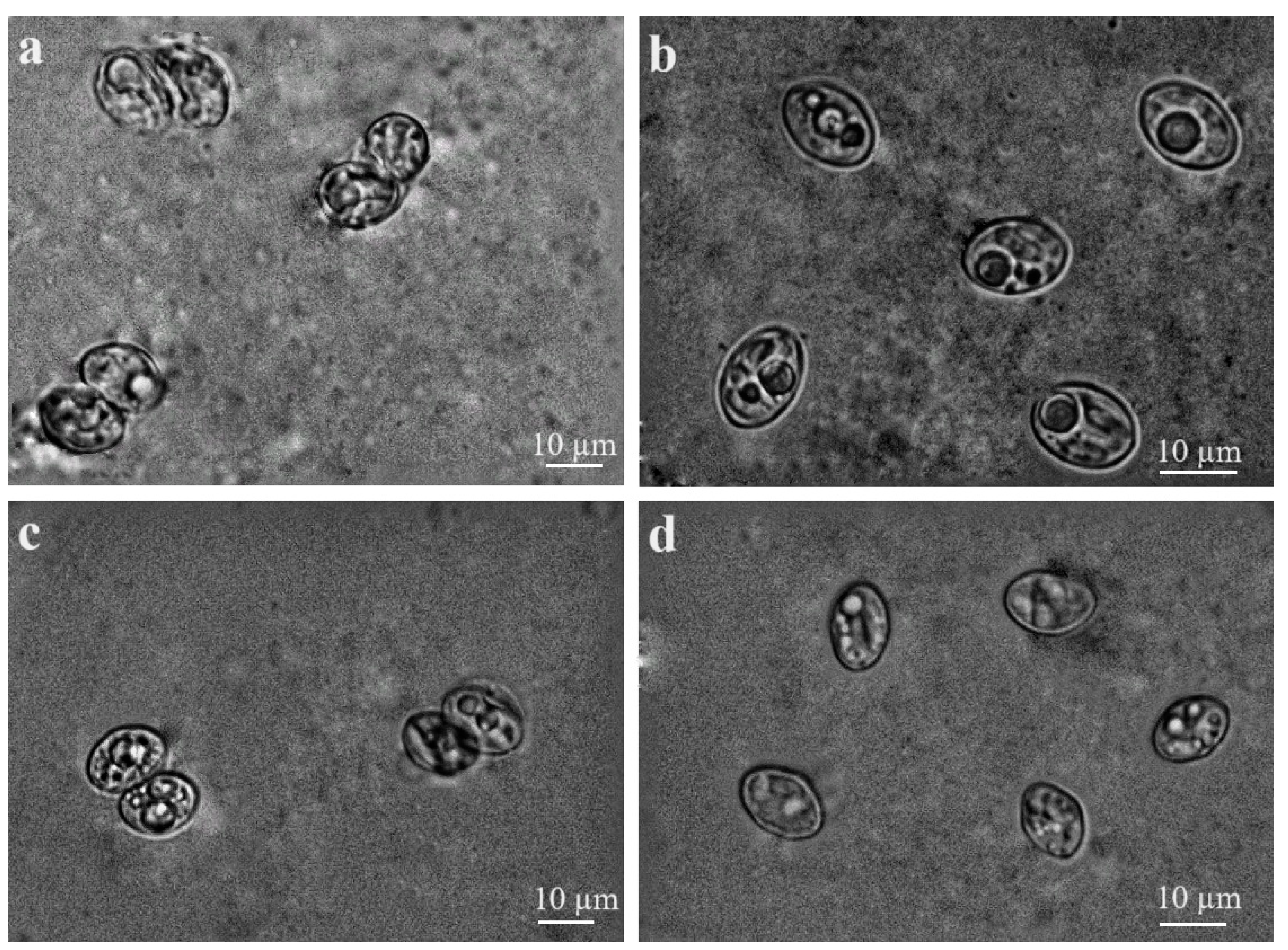
3.3. Morphological Characterisation of Sarcocystis spp. Sporocysts/Oocysts Found in Intestines of Buteo Buzzards
3.4. Molecular Investigation of Sarcocystis spp. Detected in Intestines of Buteo Hawks
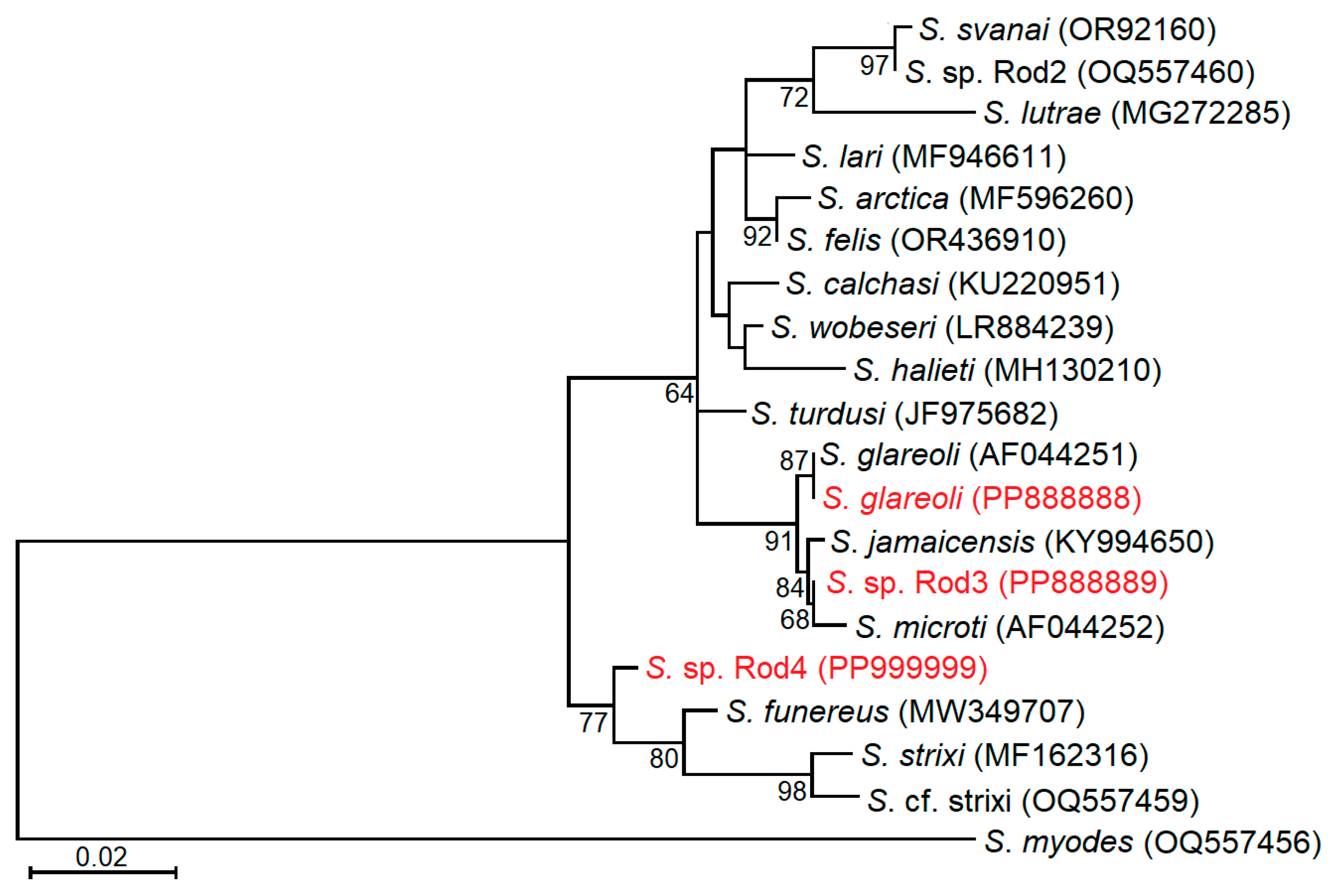
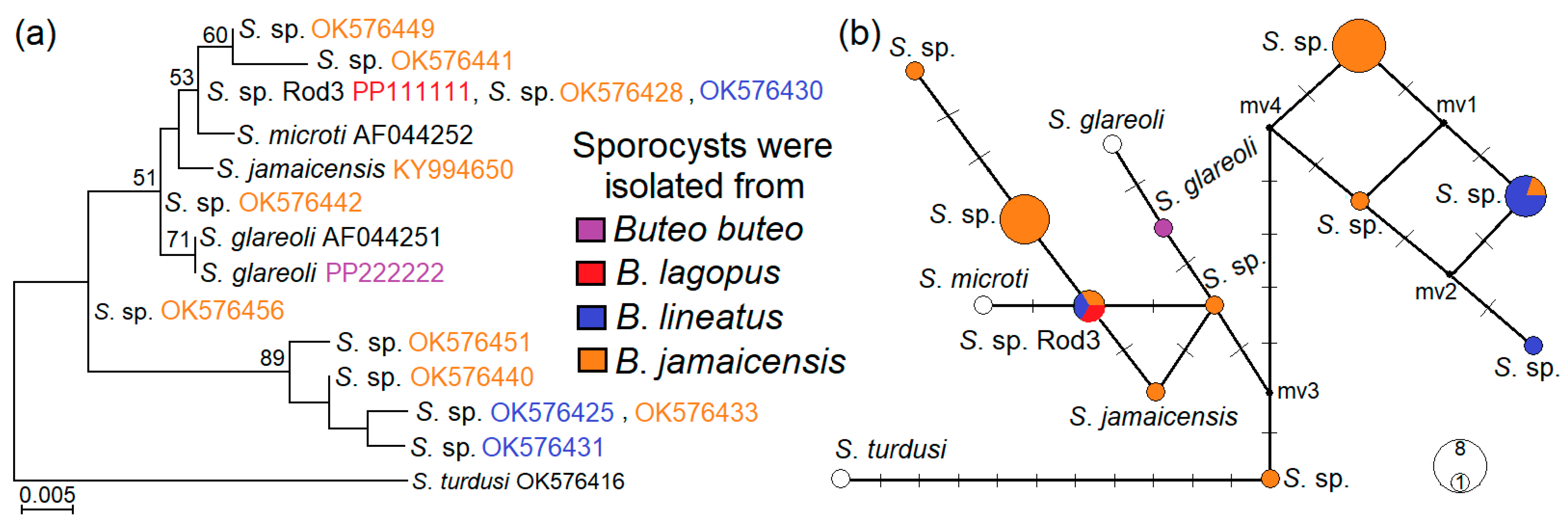
3.5. Phylogenetic Relationships of Examined Sarcocystis Species
4. Discussion
4.1. Sarcocystis Parasites in the Brain Tissues of Rodents
4.2. Buteo Buzzards as Definitive Hosts of Sarcocystis spp.
4.3. Genetic Identification of Sarcocystis sp. Rod3 and Sarcocystis sp. Rod4
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubey, J.P.; Calero-Bernal, R.; Rosenthal, B.M.; Speer, C.A.; Fayer, R. Sarcocystosis of Animals and Humans, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Gjerde, B.; Vikøren, T.; Hamnes, I.S. Molecular Identification of Sarcocystis halieti n. sp., Sarcocystis lari and Sarcocystis truncata in the Intestine of a White-Tailed Sea Eagle (Haliaeetus albicilla) in Norway. Int. J. Parasitol. Parasites Wildl. 2018, 7, 1–11. [Google Scholar] [CrossRef]
- Miller, M.A.; Barr, B.C.; Nordhausen, R.; James, E.R.; Magargal, S.L.; Murray, M.; Conrad, P.A.; Toy-Choutka, S.; Jessup, D.A.; Grigg, M.E. Ultrastructural and Molecular Confirmation of the Development of Sarcocystis neurona Tissue Cysts in the Central Nervous System of Southern Sea Otters (Enhydra lutris nereis). Int. J. Parasitol. 2009, 39, 1363–1372. [Google Scholar] [CrossRef]
- Gryz, J.; Krauze-Gryz, D. Changes in the Tawny Owl Strix aluco Diet Along an Urbanisation Gradient. Biologia 2019, 74, 279–285. [Google Scholar] [CrossRef]
- Viteri, M.C.; Stegner, M.A.; Hadly, E.A. Assessing the Reliability of Raptor Pellets in Recording Local Small Mammal Diversity. Quaternary Res. 2022, 106, 1–10. [Google Scholar] [CrossRef]
- Hu, J.; Sun, J.; Guo, Y.; Zeng, H.; Zhang, Y.; Tao, J. Infection of the Asian Gray Shrew Crocidura attenuata (Insectivora: Soricidae) with Sarcocystis attenuati n. sp. (Apicomplexa: Sarcocystidae) in China. Parasit. Vectors 2022, 15, 13. [Google Scholar] [CrossRef]
- Máca, O.; Kouba, M.; Langrová, I.; Panská, L.; Korpimäki, E.; González-Solís, D. The Tengmalm’s Owl Aegolius funereus (Aves, Strigidae) as the Definitive Host of Sarcocystis funereus sp. nov. (Apicomplexa). Front. Vet. Sci. 2024, 11, 1356549. [Google Scholar] [CrossRef]
- Jäkel, T.; Raisch, L.; Richter, S.; Wirth, M.; Birenbaum, D.; Ginting, S.; Khoprasert, Y.; Mackenstedt, U.; Wassermann, M. Morphological and Molecular Phylogenetic Characterization of Sarcocystis kani sp. nov. and Other Novel, Closely Related Sarcocystis spp. Infecting Small Mammals and Colubrid Snakes in Asia. Int. J. Parasitol. Parasites Wildl. 2023, 22, 184–198. [Google Scholar] [CrossRef]
- Qin, T.; Ortega-Perez, P.; Wibbelt, G.; Lakim, M.B.; Ginting, S.; Khoprasert, Y.; Wells, K.; Hu, J.; Jäkel, T. A Cyst-Forming Coccidian with Large Geographical Range Infecting Forest and Commensal Rodents: Sarcocystis muricoelognathis sp. nov. Parasit. Vectors 2024, 17, 135. [Google Scholar] [CrossRef]
- Rudaitytė-Lukošienė, E.; Jasiulionis, M.; Balčiauskas, L.; Prakas, P.; Stirkė, V.; Butkauskas, D. Morphological and Molecular Description of Sarcocystis myodes n. sp. from the Bank Vole (Clethrionomys glareolus) in Lithuania. Biology 2022, 11, 512. [Google Scholar] [CrossRef]
- Prakas, P.; Kirillova, V.; Gavarāne, I.; Grāvele, E.; Butkauskas, D.; Rudaitytė-Lukošienė, E.; Kirjušina, M. Morphological and Molecular Description of Sarcocystis ratti n. sp. from the Black Rat (Rattus rattus) in Latvia. Parasitol. Res. 2019, 118, 2689–2694. [Google Scholar] [CrossRef]
- Ortega Pérez, P.; Wibbelt, G.; Brinkmann, A.; Galindo Puentes, J.A.; Tuh, F.Y.Y.; Lakim, M.B.; Nitsche, A.; Wells, K.; Jäkel, T. Description of Sarcocystis scandentiborneensis sp. nov. from Treeshrews (Tupaia minor, T. tana) in Northern Borneo with Annotations on the Utility of COI and 18S rDNA Sequences for Species Delineation. Int. J. Parasitol. Parasites Wildl. 2020, 12, 220–231. [Google Scholar] [CrossRef]
- Bentancourt Rossoli, J.V.; Moré, G.; Soto-Cabrera, A.; Moore, D.P.; Morrell, E.L.; Pedrana, J.; Scioli, M.V.; Campero, L.M.; Basso, W.; Hecker, Y.P.; et al. Identification of Sarcocystis spp. in Synanthropic (Muridae) and Wild (Cricetidae) Rodents from Argentina. Parasitol. Res. 2024, 123, 31. [Google Scholar] [CrossRef]
- Geisel, O.; Kaiser, E.; Vogel, O.; Krampitz, H.E.; Rommel, M. Pathomorphologic Findings in Short-Tailed Voles (Microtus agrestis) Experimentally-Infected with Frenkelia microti. J. Wildl. Dis. 1979, 15, 267–270. [Google Scholar] [CrossRef]
- Votypka, J.; Hypša, V.; Jirku, M.; Flegr, J.; Vavra, J.; Lukes, J. Molecular Phylogenetic Relatedness of Frenkelia spp. (Protozoa, Apicomplexa) to Sarcocystis falcatula Stiles 1893: Is the Genus Sarcocystis Paraphyletic? J. Eukaryot. Microbiol. 1998, 45, 137–141. [Google Scholar] [CrossRef]
- Mugridge, N.B.; Morrison, D.A.; Johnson, A.M.; Luton, K.; Dubey, J.P.; Votýpka, J.; Tenter, A.M. Phylogenetic Relationships of the Genus Frenkelia: A Review of its History and New Knowledge Gained from Comparison of Large Subunit Ribosomal Ribonucleic Acid Gene Sequences. Int. J. Parasitol. 1999, 29, 957–972. [Google Scholar] [CrossRef]
- Mugridge, N.B.; Morrison, D.A.; Jäkel, T.; Heckeroth, A.R.; Tenter, A.M.; Johnson, A.M. Effects of Sequence Alignment and Structural Domains of Ribosomal DNA on Phylogeny Reconstruction for the Protozoan Family Sarcocystidae. Mol. Biol. Evol. 2000, 17, 1842–1853. [Google Scholar] [CrossRef]
- Modrý, D.; Votýpka, J.; Svobodová, M. Note on the Taxonomy of Frenkelia microti (Findlay & Middleton, 1934) (Apicomplexa: Sarcocystidae). Syst. Parasitol. 2004, 58, 185–187. [Google Scholar] [CrossRef]
- Odening, K. The Present State of Species-Systematics in Sarcocystis Lankester, 1882 (Protista, Sporozoa, Coccidia). Syst. Parasitol. 1998, 41, 209–233. [Google Scholar] [CrossRef]
- Laakkonen, J.; Henttonen, H. Ultrastructure of Frenkelia sp. from a Norwegian Lemming in Finland. J. Wildl. Dis. 2000, 36, 362–366. [Google Scholar] [CrossRef]
- Karstad, L. Toxoplasma Microti (The M-Organism) in the Muskrat (Ondatra Zibethica). Can. Vet. J. 1963, 4, 249–251. [Google Scholar]
- Kennedy, M.J.; Frelier, P.F. Frenkelia sp. from the Brain of a Porcupine (Erethizon Dorsatum) from Alberta, Canada. J. Wildl. Dis. 1986, 22, 112–114. [Google Scholar] [CrossRef] [PubMed]
- Fichet-Calvet, E.; Kia, E.B.; Giraudoux, P.; Quéré, J.P.; Delattre, P.; Ashford, R.W. Frenkelia Parasites in a Small Mammal Community. Dynamics of Infection and Effect on the Host. Parasite 2004, 11, 301–310. [Google Scholar] [CrossRef]
- Krücken, J.; Blümke, J.; Maaz, D.; Demeler, J.; Ramünke, S.; Antolová, D.; Schaper, R.; Von Samson-Himmelstjerna, G. Small Rodents as Paratenic or Intermediate Hosts of Carnivore Parasites in Berlin, Germany. PLoS ONE 2017, 12, e0172829. [Google Scholar] [CrossRef]
- Cabral, A.D.; Su, C.; Soares, R.M.; Gennari, S.M.; Sperança, M.A.; Da Rosa, A.R.; Pena, H.F.J. Occurrence and Diversity of Sarcocystidae Protozoa in Muscle and Brain Tissues of Bats from São Paulo State, Brazil. Int. J. Parasitol. Parasites Wildl. 2021, 14, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Upton, S.J.; McKown, R.D. The Red-Tailed Hawk, Buteo jamaicensis, a Native Definitive Host of Frenkelia microti (Apicomplexa) in North America. J. Wildl. Dis. 1992, 28, 85–90. [Google Scholar] [CrossRef]
- Laakkonen, J.; Haukisalmi, V.; Niemimaa, J.; Henttonen, H. Parasite Diversity of Norwegian Lemmings (Lemmus Lemmus). J. Zool. 2001, 253, 549–553. [Google Scholar] [CrossRef]
- Grikienienė, J.; Mažeikytė, R.; Balčiauskas, L. The First Data on Brain Parasites of the Genus Frenkelia (Protista: Coccidia) in Some Small Rodent Species in Lithuania. Acta Zoo. Litu. 2003, 13, 21–27. [Google Scholar] [CrossRef]
- Svobodová, M.; Vorisek, P.; Votypka, J.; Weidinger, K. Heteroxenous coccidia (Apicomplexa: Sarcocystidae) in the Populations of their Final and Intermediate Hosts: European Buzzard and Small Mammals. Acta Protozool. 2004, 43, 251–260. [Google Scholar]
- Fujita, O.; Oku, Y.; Ohbayashi, M. Frenkelia sp. from The Red-Backed Vole, Clethrionomys Rufocanus Bedfordiae, in Hokkaido, Japan. Jpn. J. Vet. Res. 1988, 36, 69–71. [Google Scholar]
- Toyé, P.; Tappin, N. Frenkelia glareoli: A Record from Devon. Trans R. Soc. Trop. Med. Hyg. 1972, 66, 529. [Google Scholar]
- Deter, J.; Bryja, J.; Chaval, Y.; Galan, M.; Henttonen, H.; Laakkonen, J.; Voutilainen, L.; Vapalahti, O.; Vaheri, A.; Salvador, A.R.; et al. Association between the DQA MHC Class II Gene and Puumala Virus Infection in Myodes Glareolus, the Bank Vole. Infect. Genet. Evol. 2008, 8, 450–458. [Google Scholar] [CrossRef]
- Waindok, P.; Özbakış-Beceriklisoy, G.; Janecek-Erfurth, E.; Springer, A.; Pfeffer, M.; Leschnik, M.; Strube, C. Parasites in Brains of Wild Rodents (Arvicolinae and Murinae) in the City of Leipzig, Germany. Int. J. Parasitol. Parasites Wildl. 2019, 10, 211–217. [Google Scholar] [CrossRef]
- Tadros, W.; Laarman, J.J. Current Concepts on the Biology, Evolution and Taxonomy of Tissue Cyst-Forming Eimeriid Coccidia. In Advances in Parasitology; Elsevier: Amsterdam, The Netherlands, 1982; Volume 20, pp. 293–468. ISBN 9780120317202. [Google Scholar]
- Kutkienė, L.; Prakas, P.; Sruoga, A.; Butkauskas, D. The Mallard Duck (Anas platyrhynchos) as Intermediate Host for Sarcocystis wobeseri sp. nov. from the Barnacle Goose (Branta leucopsis). Parasitol. Res. 2010, 107, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Marandykina-Prakienė, A.; Butkauskas, D.; Gudiškis, N.; Juozaitytė-Ngugu, E.; Bagdonaitė, D.L.; Kirjušina, M.; Calero-Bernal, R.; Prakas, P. Sarcocystis Species Richness in Sheep and Goats from Lithuania. Vet. Sci. 2023, 10, 520. [Google Scholar] [CrossRef]
- Prakas, P.; Stirkė, V.; Šneideris, D.; Rakauskaitė, P.; Butkauskas, D.; Balčiauskas, L. Protozoan Parasites of Sarcocystis Spp. in Rodents from Commercial Orchards. Animals 2023, 13, 2087. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Lindsay, D.S.; Grigg, M.E.; Dubey, J.P. Isolation, Culture and Cryopreservation of Sarcocystis Species. Curr. Protoc. Microbiol. 2017, 45, 20D.1.1–20D.1.27. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for General Users and for Biologist Programmers. In Bioinformatics Methods and Protocols; Humana Press: Totowa, NJ, USA, 1999; Volume 132, pp. 365–386. [Google Scholar] [CrossRef]
- Verma, S.K.; Von Dohlen, A.R.; Mowery, J.D.; Scott, D.; Rosenthal, B.M.; Dubey, J.P.; Lindsay, D.S. Sarcocystis jamaicensis n. sp., from Red-Tailed Hawks (Buteo jamaicensis) Definitive Host and IFN-γ Gene Knockout Mice as Experimental Intermediate Host. J. Parasitol. 2017, 103, 555–564. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- NETWORK v. 10.2.0.0. Available online: https://www.fluxus-engineering.com/sharenet.htm (accessed on 20 March 2024).
- Sample Size Calculators. Available online: https://sample-size.net/confidence-interval-proportion/ (accessed on 28 February 2024).
- G-Test Calculator. Available online: https://elem.com/~btilly/effective-ab-testing/g-test-calculator.html (accessed on 28 February 2024).
- Dubey, J.P.; Cerqueira-Cézar, C.K.; Murata, F.H.A.; Mowery, J.D.; Scott, D.; Dohlen, A.R.V.; Lindsay, D.S. Confirmation of Sarcocystis jamaicensis Sarcocysts in IFN-c Gene Knockout Mice Orally Inoculated with Sporocysts from a Red-Tailed Hawk (Buteo jamaicensis). J. Parasitol. 2019, 105, 143. [Google Scholar] [CrossRef]
- Verma, S.K.; Von Dohlen, A.R.; Mowery, J.D.; Scott, D.; Cerqueira-Cézar, C.K.; Rosenthal, B.M.; Dubey, J.P.; Lindsay, D.S. Sarcocystis strixi n. sp. from a Barred Owl (Strix varia) Definitive Host and Interferon Gamma Gene Knockout Mice as Experimental Intermediate Host. J. Parasitol. 2017, 103, 768–777. [Google Scholar] [CrossRef]
- Máca, O.; Kouba, M.; Korpimäki, E.; González-Solís, D. Molecular Identification of Sarcocystis sp. (Apicomplexa, Sarcocystidae) in Offspring of Tengmalm’s Owls, Aegolius funereus (Aves, Strigidae). Front. Vet. Sci. 2021, 8, 804096. [Google Scholar] [CrossRef]
- Rogers, K.H.; Arranz-Solís, D.; Saeij, J.P.; Lewis, S.; Mete, A. Sarcocystis calchasi and other Sarcocystidae detected in predatory birds in California, USA. Int. J. Parasitol. Parasites Wildl. 2022, 17, 91–99. [Google Scholar] [CrossRef]
- Hoogenboom, I.; Dijkstra, C. Sarcocystis Cernae: A Parasite Increasing the Risk of Predation of Its Intermediate Host, Microtus Arvalis. Oecologia 1987, 74, 86–92. [Google Scholar] [CrossRef]
- Rommel, M. Vergleichende Darstellung der Entwicklungsbiologie der Gattungen Sarcocystis, Frenkelia, Isospora, Cystoisospora, Hammondia, Toxoplasma und Besnoitia. Z. Parasitenkd. 1978, 57, 269–283. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Ambrus, S.I.; Blagburn, B.L. Frenkelia sp.-like infection in the small intestine of a red-tailed hawk. J. Wildl. Dis. 1987, 23, 677–679. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, D.S.; Blagburn, B.L. Caryospora uptoni and Frenkelia sp.-like Coccidial lnfections in Red-tailed Hawks (Buteo borealis). J. Wildl. Dis. 1989, 25, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Génsbøl, B. Collins Guide to the Birds of Prey of Britain and Europe, North Africa and the Middle East; HarperCollins Publishers: London, UK, 1986. [Google Scholar]
- Kontrimavičius, V.L.; Kazlauskas, R.; Logminas, V. Fauna of Lithuania. Birds; Mokslas: Vilnius, Lithuania, 1990. [Google Scholar]
- Cramp, S. (Ed.) Handbook of the Birds of Europe, the Middle East and North Africa: The Birds of the Western Palaearctic. Vol. II Hawks to Bustards; Oxford University Press: Oxford, UK, 1980; pp. 1–695. [Google Scholar]
- Reif, V.; Tornberg, R.; Jungell, S.; Korpimäki, E. Diet variation of common buzzards in Finland supports the alternative prey hypothesis. Ecography 2001, 24, 267–274. [Google Scholar] [CrossRef]
- Panek, M. Does Habitat Diversity Modify the Dietary and Reproductive Response to Prey Fluctuations in a Generalist Raptor Predator, the Eurasian Buzzard Buteo buteo? Birds 2021, 2, 114–126. [Google Scholar] [CrossRef]
- Prakas, P.; Butkauskas, D.; Švažas, S.; Juozaitytė-Ngugu, E.; Stanevičius, V. Morphologic and Genetic Identification of Sarcocystis fulicae n. sp. (Apicomplexa: Sarcocystidae) from the Eurasian coot (Fulica atra). J. Wildl. Dis. 2018, 54, 765–771. [Google Scholar] [CrossRef]
- Máca, O. Molecular Identification of Sarcocystis lutrae (Apicomplexa: Sarcocystidae) from the Raccoon Dog, Nyctereutes procyonoides, and the Common Raccoon, Procyon lotor, in the Czech Republic. Parasit. Vectors 2020, 13, 231. [Google Scholar] [CrossRef]
- Prakas, P.; Gudiškis, N.; Kitrytė, N.; Bagdonaitė, D.L.; Baltrūnaitė, L. Detection of Three Sarcocystis Species (Apicomplexa) in Blood Samples of the Bank Vole and Yellow-Necked Mouse from Lithuania. Life 2024, 14, 365. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Orientation | Primer Sequence | Type of PCR | Ta | LP | Reference |
|---|---|---|---|---|---|---|
| KL-P1F | Forward | TACCCGCTGAACTTAAGCAT | conventional | 52 | ~1000 | [35] |
| KL-P1R | Reverse | CCCAAGTTTGACGAACGATT | conventional | |||
| Sgrau281 | Forward | GAACAGGGAAGAGCTCAAAGTG | nested | 63 | ~900 | [37] |
| Sgrau282 | Reverse | GGTTTCCCCTGACTTCATTCTAC | nested | |||
| GsSglaF1 | Forward | GCAAAATGTGTGGTAAGTTTCACAT | nested | 61 | ~560 | Present study |
| GsSglaR1 | Reverse | CCCTCTAAAAAGATGTTACCCTTCT | nested | |||
| GsSmicF1 | Forward | TGTGGTAAGTTTCACATAAGGCTAA | nested | 61 | ~550 | |
| GsSmicR1 | Reverse | CTTTCTAAAAAGATGTACCTTCTCCT | nested |
| Investigation Sites | DR, % | DR CI | Species | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. agr | A. fla | C. gla | M. agr | M. arv | A. oec | M. min | M. mus | N. fod | S. ara | S. min | |||
| 1. Aukštikalniai | 16.7 | 0.4–64.1 | 6 (1) | 7 (0) | |||||||||
| 2. Bileišiai | 10.0 | 5.7–15.9 | 1 (0) | 150 (15) | 5 (0) | 16 (0) | 1 (0) | 1 (0) | 16 (0) | 31 (0) | |||
| 3. Juodkrantė | 9 (0) | ||||||||||||
| 4. Kamasta | 22.5 | 10.8–38.5 | 40 (9) | 1 (0) | 3 (0) | 10 (0) | 8 (0) | ||||||
| 5. Kukinis | 12.5 | 0.3–52.6 | 8 (1) | 2 (0) | 5 (0) | 3 (0) | 1 (0) | ||||||
| 6. Lukštas | 2.1 | 0.1–11.3 | 1 (0) | 47 (1) | 1 (0) | 7 (0) | 2 (0) | 1 (0) | 2 (0) | 3 (0) | |||
| 7. Mieliūnai | 5 (0) | 2 (0) | 53 (0) | 4 (0) | 21 (0) | 3 (0) | |||||||
| 8. Sudervė | 24 (0) | 21 (0) | 1 (0) | 5 (0) | 6 (0) | 4 (0) | 1 (0) | ||||||
| 9. Šešuolėliai | 7.7 | 0.2–36.0 | 13 (1) | ||||||||||
| 10. Utena | 7.6 | 2.5–16.8 | 66 (5) | 4 (0) | 30 (0) | 5 (0) | 1 (0) | 2 (0) | 1 (0) | 7 (0) | 10 (0) | ||
| 11. Vilnius | 5 (0) | ||||||||||||
| 12. Zabarauskai | 20.0 | 0.5–71.6 | 5 (1) | 1 (0) | 1 (0) | ||||||||
| 13. Žiežmariai | 4 (0) | 1 (0) | |||||||||||
| Total: | 5 (0) | 2 (0) | 374 (34) | 14 (0) | 144 (0) | 11 (0) | 3 (0) | 33 (0) | 9 (0) | 45 (0) | 54 (0) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakas, P.; Jasiulionis, M.; Šukytė, T.; Juozaitytė-Ngugu, E.; Stirkė, V.; Balčiauskas, L.; Butkauskas, D. First Observations of Buzzards (Buteo) as Definitive Hosts of Sarcocystis Parasites Forming Cysts in the Brain Tissues of Rodents in Lithuania. Biology 2024, 13, 264. https://doi.org/10.3390/biology13040264
Prakas P, Jasiulionis M, Šukytė T, Juozaitytė-Ngugu E, Stirkė V, Balčiauskas L, Butkauskas D. First Observations of Buzzards (Buteo) as Definitive Hosts of Sarcocystis Parasites Forming Cysts in the Brain Tissues of Rodents in Lithuania. Biology. 2024; 13(4):264. https://doi.org/10.3390/biology13040264
Chicago/Turabian StylePrakas, Petras, Marius Jasiulionis, Tautvilė Šukytė, Evelina Juozaitytė-Ngugu, Vitalijus Stirkė, Linas Balčiauskas, and Dalius Butkauskas. 2024. "First Observations of Buzzards (Buteo) as Definitive Hosts of Sarcocystis Parasites Forming Cysts in the Brain Tissues of Rodents in Lithuania" Biology 13, no. 4: 264. https://doi.org/10.3390/biology13040264








