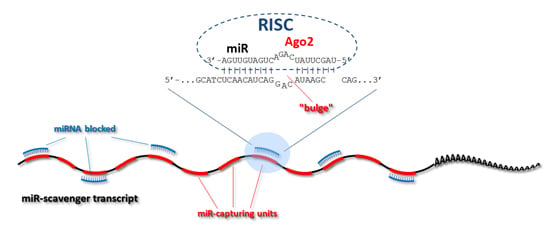
A Versatile Tool for Stable Inhibition of microRNA Activity
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. Design of the Oligos
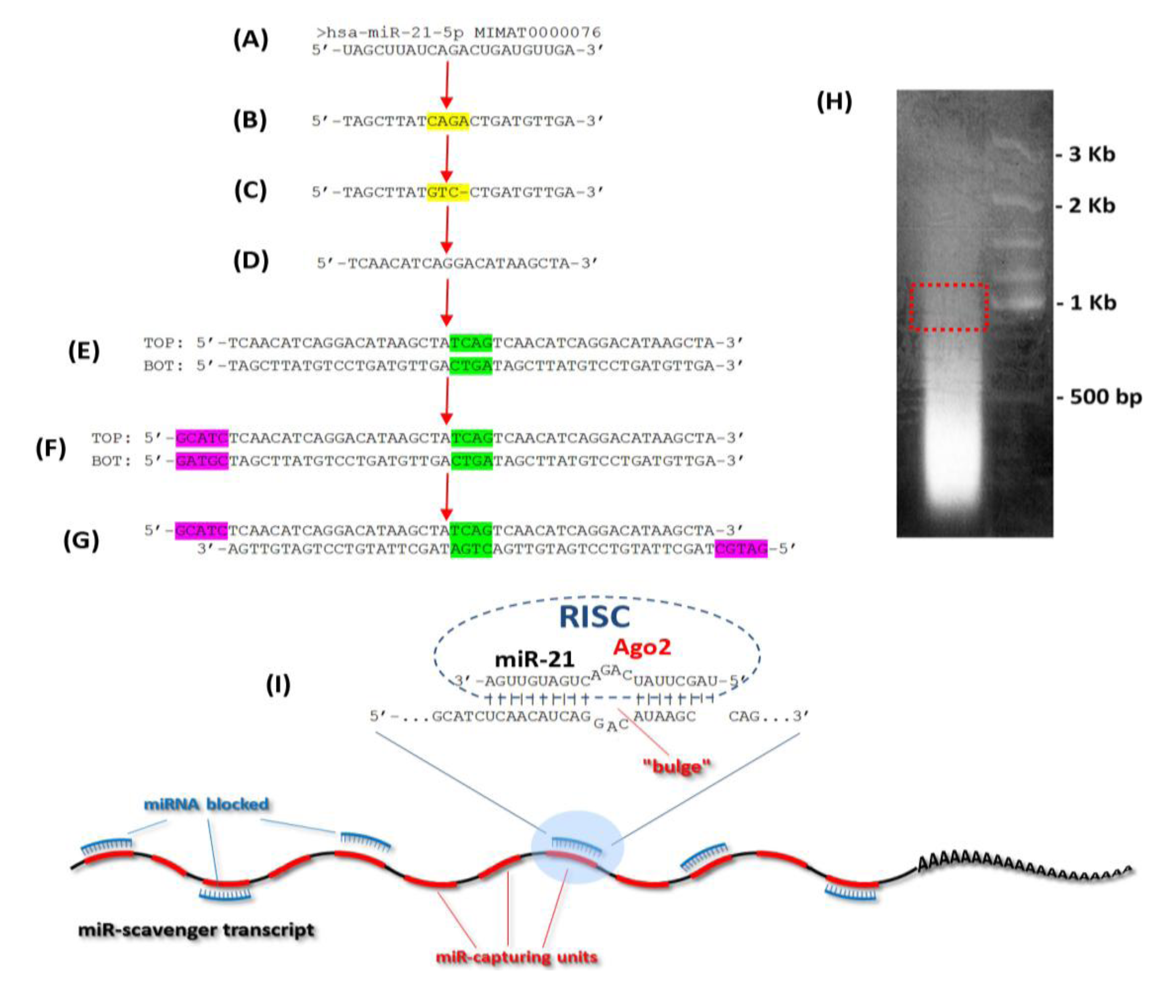

3.2. Concatemerization Step
3.3. Cloning of the Concatemerized miR Scavenger Sequence
3.4. Lentivirus Production
3.5. Transduction of LA7 or mESC Cells
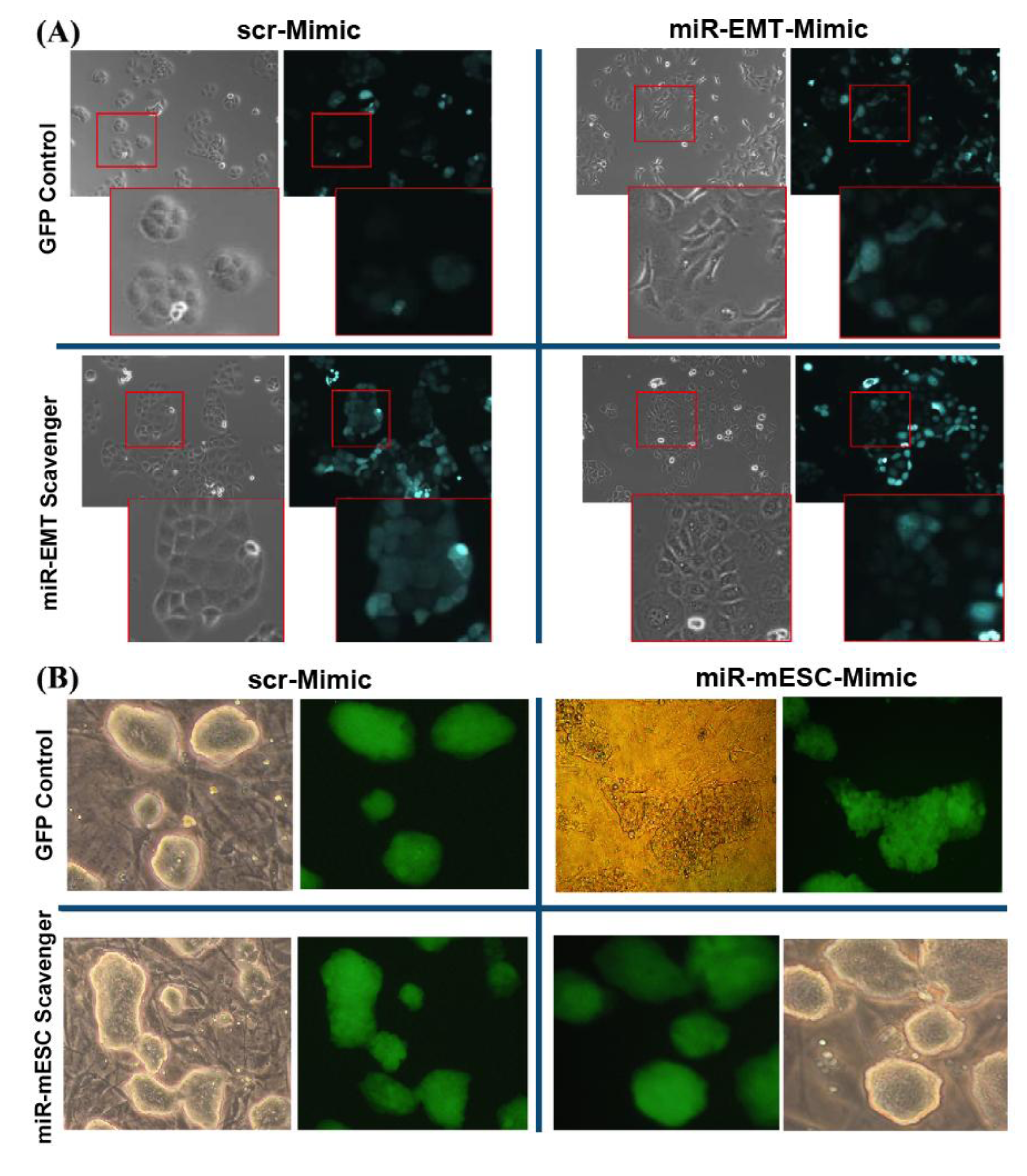
3.6. Inhibition of Mimic Oligo miR Activity by the miR Scavengers
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar]
- Tsang, J.S.; Ebert, M.S.; van Oudenaarden, A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol. Cell 2010, 38, 140–153. [Google Scholar] [CrossRef] [Green Version]
- Mukherji, S.; Ebert, M.S.; Zheng, G.X.; Tsang, J.S.; Sharp, P.A.; van Oudenaarden, A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011, 43, 854–859. [Google Scholar] [Green Version]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar]
- Carè, A.; Catalucci, D.; Felicetti, F.; Bonci, D.; Addario, A.; Gallo, P.; Bang, M.L.; Segnalini, P.; Gu, Y.; Dalton, N.D.; et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007, 13, 613–618. [Google Scholar] [CrossRef]
- Gentner, B.; Schira, G.; Giustacchini, A.; Amendola, M.; Brown, B.D.; Ponzoni, M.; Naldini, L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods 2009, 6, 63–66. [Google Scholar] [CrossRef]
- Sayed, D.; Rane, S.; Lypowy, J.; He, M.; Chen, I.Y.; Vashistha, H.; Yan, L.; Malhotra, A.; Vatner, D.; Abdellatif, M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell 2008, 19, 3272–3282. [Google Scholar] [CrossRef]
- Scherr, M.; Venturini, L.; Battmer, K.; Schaller-Schoenitz, M.; Schaefer, D.; Dallmann, I.; Ganser, A.; Eder, M. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007, 35, e149. [Google Scholar] [CrossRef]
- Haraguchi, T.; Ozaki, Y.; Iba, H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009, 37, e43. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Korkola, J.E.; Sadelain, M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells 2010, 28, 287–296. [Google Scholar]
- Huang, J.; Zhao, L.; Xing, L.; Chen, D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 2010, 28, 357–364. [Google Scholar]
- Zucchi, I.; Sanzone, S.; Astigiano, S.; Pelucchi, P.; Scotti, M.; Valsecchi, V.; Barbieri, O.; Bertoli, G.; Albertini, A.; Reinbold, R.A.; et al. The properties of a mammary gland cancer stem cell. Proc. Natl. Acad. Sci. USA 2007, 104, 10476–10481. [Google Scholar]
- Zucchi, I.; Astigiano, S.; Bertalot, G.; Sanzone, S.; Cocola, C.; Pelucchi, P.; Bertoli, G.; Stehling, M.; Barbieri, O.; Albertini, A.; et al. Distinct populations of tumor-initiating cells derived from a tumor generated by rat mammary cancer stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16940–16945. [Google Scholar] [CrossRef]
- Tsang, J.; Zhu, J.; van Oudenaarden, A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 2007, 26, 753–767. [Google Scholar] [CrossRef]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef]
- Leone, V.; D’Angelo, D.; Ferraro, A.; Pallante, P.; Rubio, I.; Santoro, M.; Croce, C.M.; Fusco, A. A TSH-CREB1-microRNA loop is required for thyroid cell growth. Mol. Endocrinol. 2011, 25, 1819–1830. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Zhang, H.; Chen, Y. MicroRNA-mediated positive feedback loop and optimized bistable switch in a cancer network Involving miR-17-92. PLoS One 2011, 6, e26302. [Google Scholar]
- Johnston, R.J., Jr.; Chang, S.; Etchberger, J.F.; Ortiz, C.O.; Hobert, O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA 2005, 102, 12449–12454. [Google Scholar]
- miRBase: The microRNA Database. Available online: http://www.mirbase.org/ (accessed on 23 May 2013).
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar]
- Ghelani, H.S.; Rachchh, M.A.; Gokani, R.H. MicroRNAs as newer therapeutic targets: A big hope from a tiny player. J. Pharmacol. Pharmacother. 2012, 3, 217–227. [Google Scholar]
- Lindow, M.; Kauppinen, S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012, 199, 407–412. [Google Scholar]
- Alvarez, M.L.; Distefano, J.K. Towards microRNA-based therapeutics for diabetic nephropathy. Diabetologia 2013, 56, 444–456. [Google Scholar] [CrossRef]
- Nikaki, A.; Piperi, C.; Papavassiliou, A.G. Role of microRNAs in gliomagenesis: Targeting miRNAs in glioblastoma multiforme therapy. Exp. Opin. Invest. Drugs 2012, 21, 1475–1488. [Google Scholar]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar]
- Singh, R.; Mo, Y.Y. Role of microRNAs in breast cancer. Cancer Biol. Ther. 2013, 14, 201–212. [Google Scholar]
- Cho, W.C. Exploiting the therapeutic potential of microRNAs in human cancer. Expert Opin. Ther. Targets 2012, 16, 345–350. [Google Scholar]
- Nana-Sinkam, S.P.; Croce, C.M. Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther. 2013, 93, 98–104. [Google Scholar]
- Bonci, D.; Coppola, V.; Musumeci, M.; Addario, A.; Giuffrida, R.; Memeo, L.; D’Urso, L.; Pagliuca, A.; Biffoni, M.; Labbaye, C.; et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008, 14, 1271–1277. [Google Scholar]
- Starczynowski, D.T.; Kuchenbauer, F.; Argiropoulos, B.; Sung, S.; Morin, R.; Muranyi, A.; Hirst, M.; Hogge, D.; Marra, M.; Wells, R.A.; et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 2010, 16, 49–58. [Google Scholar]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009, 10, 1252–1259. [Google Scholar] [CrossRef]
- Loya, C.M.; Lu, C.S.; van Vactor, D.; Fulga, T.A. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods 2009, 6, 897–903. [Google Scholar]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar]
- Rasmussen, K.D.; Simmini, S.; Abreu-Goodger, C.; Bartonicek, N.; di Giacomo, M.; Bilbao-Cortes, D.; Horos, R.; von Lindern, M.; Enright, A.J.; O’Carroll, D. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 2010, 207, 1351–1358. [Google Scholar]
- Hübner, K.; Fuhrmann, G.; Christenson, L.K.; Kehler, J.; Reinbold, R.; de la Fuente, R.; Wood, J.; Strauss, J.F., III; Boiani, M.; Schöler, H.R. Derivation of oocytes from mouse embryonic stem cells. Science 2003, 300, 1251–1256. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pelucchi, P.; Tria, V.; Martino, V.; Sabour, D.; Bertalot, G.; Molgora, S.; Palizban, M.; Götte, M.; Zucchi, I.; Reinbold, R.A. A Versatile Tool for Stable Inhibition of microRNA Activity. Biology 2013, 2, 861-871. https://doi.org/10.3390/biology2030861
Pelucchi P, Tria V, Martino V, Sabour D, Bertalot G, Molgora S, Palizban M, Götte M, Zucchi I, Reinbold RA. A Versatile Tool for Stable Inhibition of microRNA Activity. Biology. 2013; 2(3):861-871. https://doi.org/10.3390/biology2030861
Chicago/Turabian StylePelucchi, Paride, Valeria Tria, Valentina Martino, Davood Sabour, Giovanni Bertalot, Stefano Molgora, Mira Palizban, Martin Götte, Ileana Zucchi, and Rolland A. Reinbold. 2013. "A Versatile Tool for Stable Inhibition of microRNA Activity" Biology 2, no. 3: 861-871. https://doi.org/10.3390/biology2030861
APA StylePelucchi, P., Tria, V., Martino, V., Sabour, D., Bertalot, G., Molgora, S., Palizban, M., Götte, M., Zucchi, I., & Reinbold, R. A. (2013). A Versatile Tool for Stable Inhibition of microRNA Activity. Biology, 2(3), 861-871. https://doi.org/10.3390/biology2030861