Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Expression and Purification of Two-Domain Adhesin Lectin Domains
2.2. CFG Glycan Microarray Screening
2.3. Synthesis of a GlcNAcβ1-xGal Disaccharide Series
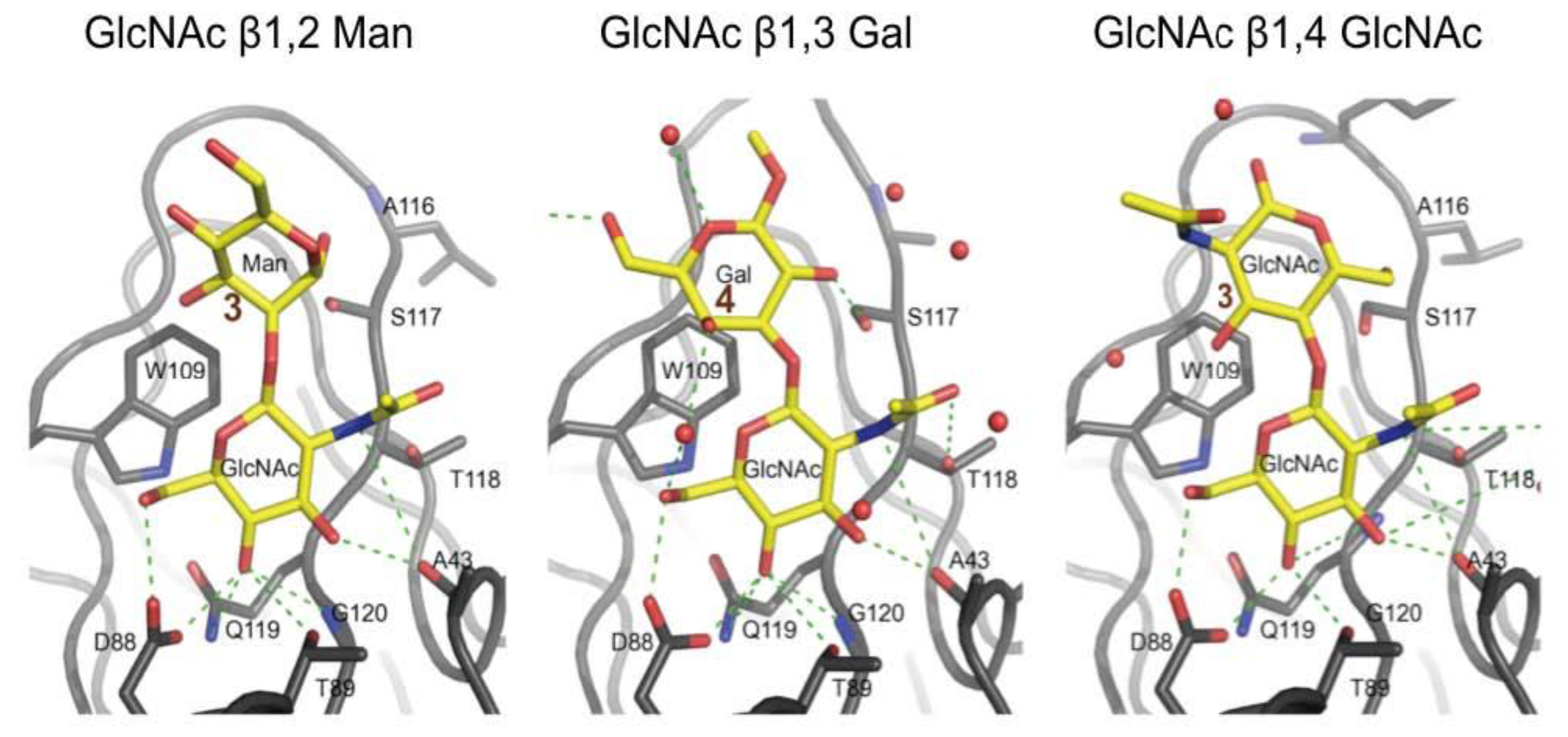
2.4. Surface Plasmon Resonance Measurements for Linkage Discrimination by F17G
2.5. Shotgun Glycan Microarray Production
2.6. Natural and Shotgun Glycan Microarrays Screening
2.7. Crystallization and Structure Solution
3. Results
3.1. Synthetic and Natural Glycan Microarray Screening
| N° | Glycan name | F17aG | F17bG | F17dG | F17eG | F17fG |
|---|---|---|---|---|---|---|
| 167 | GlcNAcβ1,3Galβ1,4Glcβ-Sp0 | 57,013 (5,191) | 13,149 (2,286) | 29,181 (3,281) | 54,005 (5,715) | 26,336 (7,255) |
| 165 | GlcNAcβ1-3Galβ1,4GlcNAcβ-Sp8 | 51,880 (5,023) | 15,483 (1,729) | 17,991 (4,401) | 49,223 (5,377) | 28,888 (6,925) |
| 160 | GlcNAcβ1,3(GlcNAcβ1,6)Galβ1,4GlcNAcβ-Sp8 | 44,391 (3,949) | 21,630 (2,490) | 17,195 (3,442) | 41,907 (4,694) | 37,083 (5,973) |
| 159 | GlcNAcβ1,3(GlcNAcβ1,6)GalNAcα-Sp8 | 35,858 (4,079) | 12,183 (1,524) | 23,756 (2,105) | 46,840 (6,520) | 37,083 (1,197) |
| 162 | GlcNAcβ1,3Galβ-Sp8 | 32,288 (4,175) | 16,310 (1,907) | 16,277 (9,959) | 38,836 (5,631) | 27,648 (1,137) |
| 161 | GlcNAcβ1,3GalNAcα-Sp8 | 25,971 (3,174) | 15,182 (2,925) | 6,635 (2,735) | 33,330 (7,321) | 10,999 (2,929) |
| 88 | GalNAcβ1,3GalNAcα-Sp8 | 24,610 (3,462) | 17,169 (4,228) | 11,034 (1204) | 50,791 (5,749) | 10,691 (3,009) |
| 25 | (GlcNAcβ1,3(GlcNAcβ1,6)GlcNAcβ1,4)GlcNAc-Sp8 | 20,305 (2,023) | 6,148 (1,367) | 8,080 (1,518) | 50,775 (6,375) | 2,992 (992) |
| 21 | GlcNAcβ-Sp0 | 15,621 (2,697) | 14,972 (2,705) | 3,377 (643) | 34,825 (6,475) | 5,354 (1,572) |
| 22 | GlcNAcβ-Sp8 | 22,909 (3,087) | 12,289 (1,168) | 2,514 (365) | 31,863 (4,615) | 10,738 (2,670) |
| 174 | GlcNAcβ1,6(Galβ1,3)GalNAcα-Sp8 | 12,541 (1,115) | 6,017 (587) | 3,492 (658) | 46,930 5,852 | 3,206(997) |
| 170 | GlcNAcβ1,4Galβ1,4GlcNAcβ-Sp8 | 12,499 (1,921) | 13,001 (2,039) | 1,056 (190) | 39,794 (6,245) | 7,176 (2,338) |
| 175 | GlcNAcβ1,6GalNAcα-Sp8 | 11,120 (1,871) | 6,683 (947) | 2,432 (406) | 43,848 (4,140) | 26,522 (11,365) |
| 166 | GlcNAcβ1,3Galβ1,4GlcNAcβ1,3Galβ1,4GlcNAcβ-Sp0 | 10,917 (811) | 7,563 (2,744) | 3,599 (772) | 43,425 (4,065) | 4,334 (856) |
| 121 | Galβ1,3(GlcNAcβ1,6)GalNAcα-Sp8 | 10,256 (1,410) | 6,215 (1,046) | 3,850 (364) | 50,893 (5,540) | 3,195 (811) |
| 176 | GlcNAcβ1,6Galβ1,4GlcNAcβ-Sp8 | 9,029 (1,096) | 5,372 (746) | 5,119 (1,633) | 40,973 (6,555) | 2,269 (561) |
| 163 | GlcNAcβ1,3Galβ1,3GalNAcα-Sp8 | 7,713 (866) | 10,655 (880) | 3,999 (1,481) | 47,984 (3,178) | 31,661 (2,807) |
| 169 | GlcNAcβ1,4(GlcNAcβ1,6)GalNAcα-Sp8 | 4,520 (2,808) | 12,960 (1,692) | 1,680 (92) | 38,200 (2,394) | 19,830 (4,279) |
| 158 | GlcNAcβ1,2Galβ1,3GalNAcα-Sp8 | 5,082 (4,924) | 1,016 (239) | 75 (35) | 305 (148) | 563 (125) |
| 23 | GlcNH2β-Sp8 | 554 (117) | 7,281 (1,506) | 1,950 (1,358) | 548 (256) | 1,385 (464) |
| 152 | Galβ1,4GlcNAcβ-Sp0 | 1,703 (1,070) | 1,974 (289) | 249 (94) | 19,659 (4,594) | 1,279 (106) |
| 173 | GlcNAcβ1,4GlcNAcβ1,4GlcNAcβ-Sp8 | 848 (88) | 2,860 (706) | 1,690 (251) | 5,689 (1,085) | 2,418 (613) |
| 148 | Galβ1,4GlcNAcβ1,3Galβ1,4Glcβ-Sp0 | 391 (24) | 1,090 (262) | 919 (375) | 3,942 (857) | 663 (146) |
| 213 | Neu5Acα2,3(Neu5Acα2,6)GalNAcα-Sp8 | 4,527 (4,043) | 4,331 (1,760) | 1,388 (1,162) | 1,229 (1,162) | 1,717 (229) |

3.2. Validation of the Glycan Array Using Surface Plasmon Resonance
| Carbohydrate | F17aG | F17bG | F17fG | F17eG |
|---|---|---|---|---|
| GlcNAcβ1,3Gal | +++ 0.66 | ++ | +++ | +++ 0.28 |
| GlcNAcβ1,4GlcNAc (chitobiose) | ++ 3.3 | ++ | ++ | |
| GlcNAc ( N-acetylglucosamine) | ++ 1.2 | ++ | ++ | |
| Me β N-acetylglucosamine | ++ 1.2 | |||
| Me α N-acetylglucosamine | + 9.8 | |||
| GlcNAcβ1,2Gal | ++ 4.2 | ++ 2.0 | ||
| GlcNAcβ1,4Gal | ++ 3.3 | ++ 2.7 | ||
| GlcNAcβ1,6Gal | ++ 2.8 | ++ 3.8 | ||
| GlcNAcβ1,2Manα1,6(GlcNAcβ1,2Manα1,3)Man | ++ 3.0 | ++ 3.0 | ||
| Gal β1,4 GlcNAc | ++ | ++ | ||
| Galβ1,6GlcNAc | + | + | + | ++ |
| Galβ1,3GlcNAcβ 1,3Galβ1,4Glc | - | |||
| Galβ1,4GlcNAcβ 1,3Gal β1,4Glc | + | |||
| Fucα1,2Galβ1,3GlcNAcβ1,3Galβ1,4Glc | - | |||
| Neu5Acα2,6Galβ1,4Glc | ++ | |||
| pNP-GlcNAc * | +++ | + | ++ | |
| N-acetylmannosamine | ++ | + | ++ | |
| N-acetylneuraminic acid | - | - | - | |
| Galβ1,3GalNAc | + | + | ||
| GlcNAc6SO3− | + | - | ++ | |
| GlcNAc3SO3− | ++ | ++ | - | |
| GlcNAc6PO32− | - | + | - | |
| GlcNH2-MurNAc * | + | + | - | |
| Neu5Acα2,3Galβ1,3(Neu5Acα2,6)GlcNAcβ1,3Gal β1,4Glc | + | + | - | |
| LeY | - | - | ++ | |
| GM1 ganglioside * | - | + | - | |
| Neu5Acα2,3Galβ1,4GlcNAcβ1,2 Man | + | + | ||
| GalNAcβ1,4GlcNAcβ1-methoxy phenyl | ++ |
3.3. Co-Crystals of F17G Variants with Disaccharides

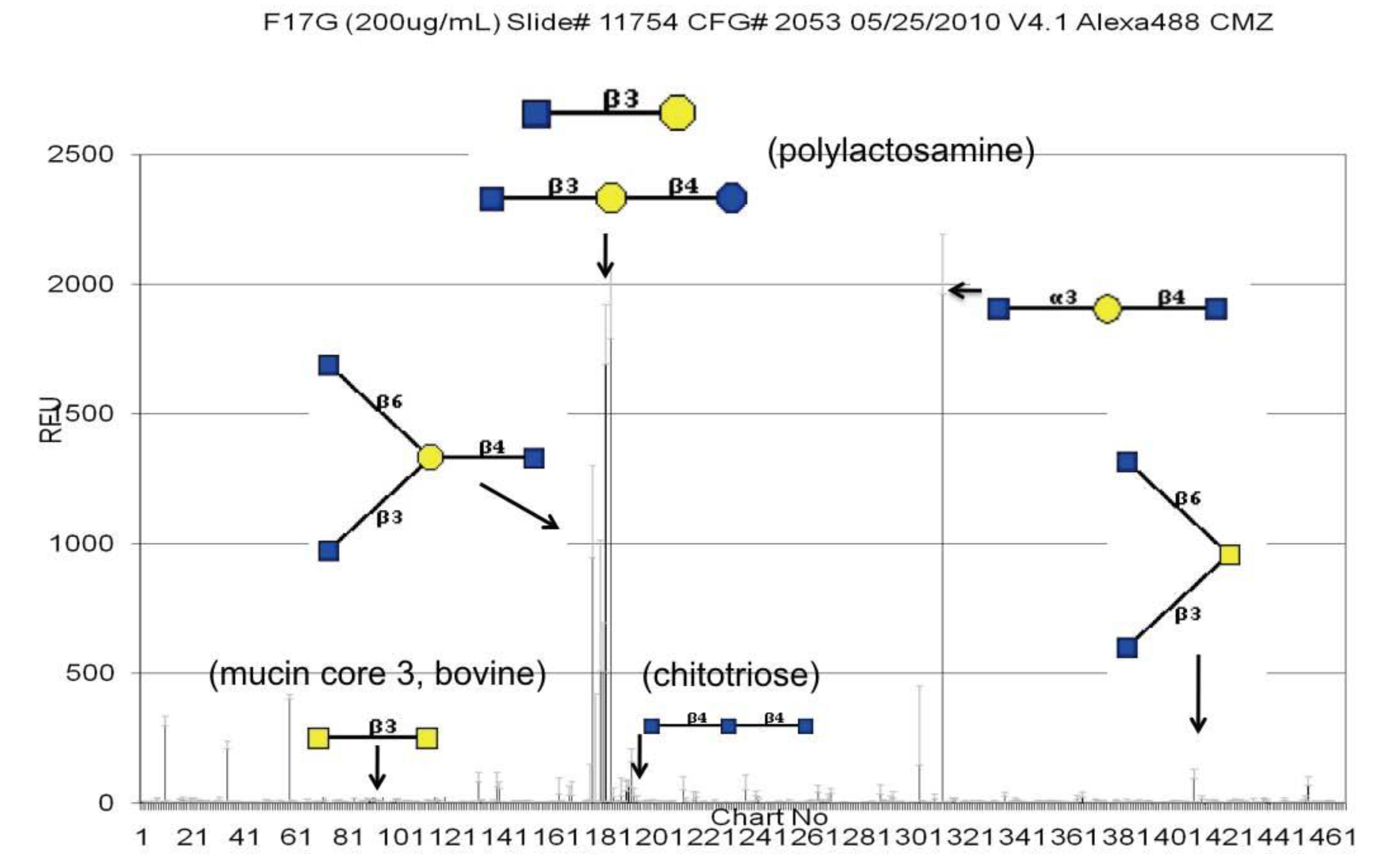
3.4. Shotgun Glycan Arrays for FedF and Whole F18-Fimbriated E. coli

3.5. F17G Variants Bind Acids and Sulfates on the Glycan Array Structures and in the Crystal
3.6. FedF Binding to Blood Group Type 1 Core Glycans with Attraction to Acid Glycosphigolipids
| candidate sulphated lactose structures | RFU | Stdev | SEM | |
|---|---|---|---|---|
| 1 | [3OSO3][6OSO3]Galβ1,4[6OSO3]GlcNAc | 10,388 | 3,852 | 1,573 |
| 2 | [3OSO3]Galβ1,4[6OSO3]GlcNAc | 6,806 | 1,854 | 757 |
| 3 | [6OSO3]Galβ1,4[6OSO3]Glc | 5,405 | 2,430 | 992 |
| 4 | [3OSO3][6OSO3]Galβ1,4GlcNAc | 4,733 | 1,157 | 472 |
| 5 | [4OSO3][6OSO3]Galβ1,4GlcNAc | 4,568 | 2,057 | 840 |
| 6 | [3OSO3]Galβ1,4[6OSO3]Glc | 4,073 | 1,515 | 618 |
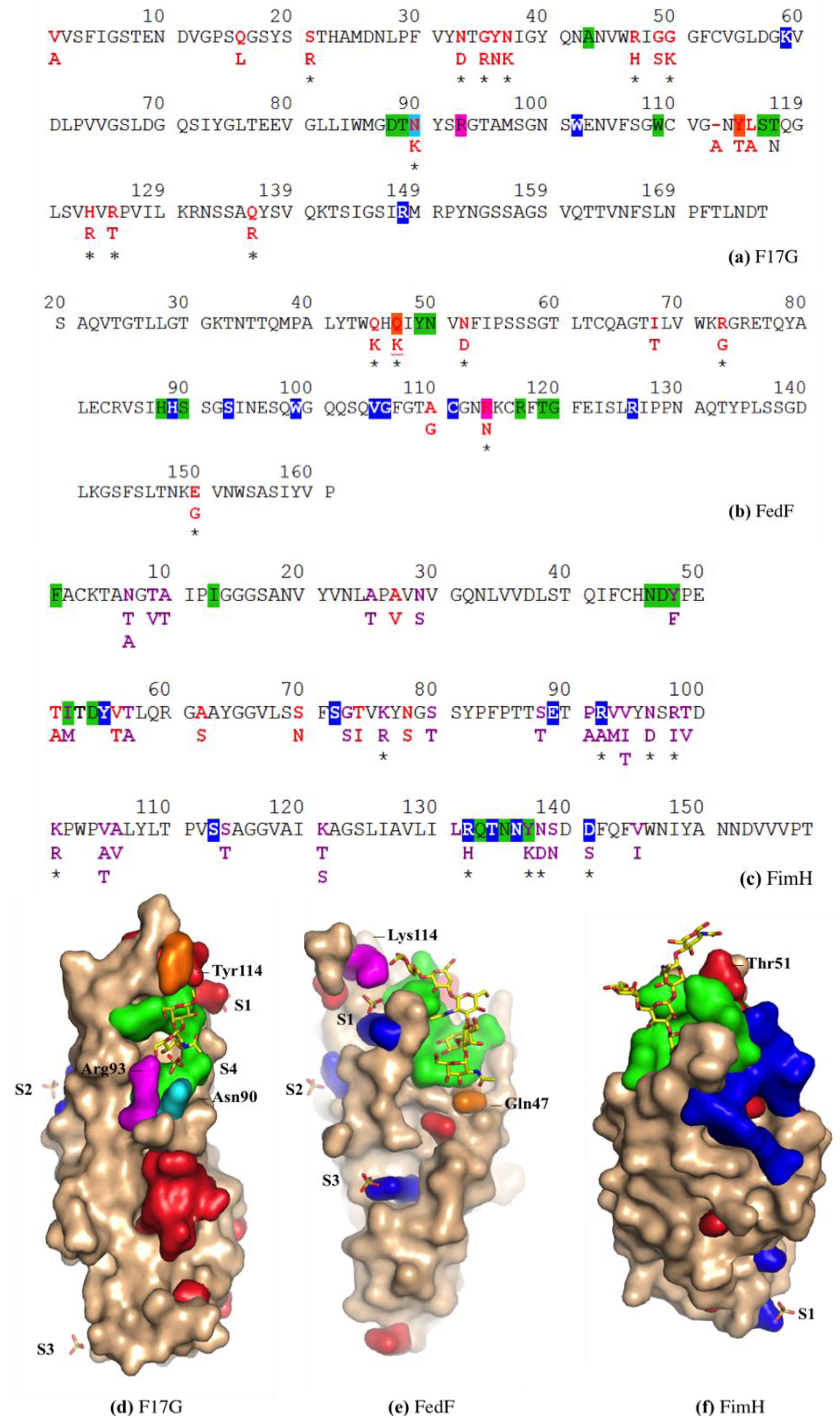
3.7. FimH Has a Charged Cradle over the Edge on the other Side of Its Oligomannoside Binding Site
4. Discussion
Acknowledgements
Conflict of Interest
References
- Kapitany, R.A.; Forsyth, G.W.; Scoot, A.; Mckenzie, S.F.; Worthington, R.W. Isolation and Partial Characterization of 2 Different Heat-Stable Enterotoxins Produced by Bovine and Porcine Strains of Enterotoxigenic Escherichia coli. Infect. Immun. 1979, 26, 173–177. [Google Scholar]
- Lintermans, P.F.; Pohl, P.; Bertels, A.; Charlier, G.; Vandekerckhove, J.; van Damme, J.; Shoup, J.; Schlicker, C.; Korhonen, T.; de Greve, H.; et al. Characterization and purification of the F17 adhesin on the surface of bovine enteropathogenic and septicemic Escherichia coli. Am. J. Vet. Res. 1988, 49, 1794–1799. [Google Scholar]
- Hahn, E.; Wild, P.; Schraner, E.M.; Bertschinger, H.U.; Haner, M.; Muller, S.A.; Aebi, U. Structural analysis of F18 fimbriae expressed by porcine toxigenic Escherichia coli. J. Struct. Biol. 2000, 132, 241–250. [Google Scholar] [CrossRef]
- Choudhury, D.; Thompson, A.; Stojanoff, V.; Langermann, S.; Pinkner, J.; Hultgren, S.J.; Knight, S.D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 1999, 285, 1061–1066. [Google Scholar] [CrossRef]
- Lintermans, P.; Pohl, P.; Deboeck, F.; Bertels, A.; Schlicker, C.; Vandekerckhove, J.; Vandamme, J.; Vanmontagu, M.; DeGreve, H. Isolation and Nucleotide-Sequence of the F17-A Gene Encoding the Structural Protein of the F-17 Fimbriae in Bovine Entero-Toxigenic Escherichia coli. Infect. Immun. 1988, 56, 1475–1484. [Google Scholar]
- Lintermans, P.F.; Bertels, A.; Schlicker, C.; Deboeck, F.; Charlier, G.; Pohl, P.; Norgren, M.; Normark, S.; van Montagu, M.; de Greve, H. Identification, characterization, and nucleotide sequence of the F17-G gene, which determines receptor binding of Escherichia coli F17 fimbriae. J. Bacteriol. 1991, 173, 3366–3373. [Google Scholar]
- Smeds, A.; Hemmann, K.; Jakava-Viljanen, M.; Pelkonen, S.; Imberechts, H.; Palva, A. Characterization of the adhesin of Escherichia coli F18 fimbriae. Infect. Immun. 2001, 69, 7941–7945. [Google Scholar] [CrossRef]
- Knight, S.D.; Bouckaert, J. Structure, function, and assembly of Type 1 fimbriae. Top. Curr. Chem. 2009, 288, 67–107. [Google Scholar] [CrossRef]
- Westerlund-Wikström, B.; Korhonen, T.K. Molecular structure of adhesin domains in Escherichia coli fimbriae. Int. J. Med. Microbiol. 2005, 295, 479–486. [Google Scholar] [CrossRef]
- De Greve, H.; Wyns, L.; Bouckaert, J. Combining sites of bacterial fimbriae. Curr. Opin. Struct. Biol. 2007, 17, 506–512. [Google Scholar] [CrossRef]
- Bouckaert, J.; Mackenzie, J.; de Paz, J.L.; Chipwaza, B.; Choudhury, D.; Zavialov, A.; Mannerstedt, K.; Anderson, J.; Pierard, D.; Wyns, L.; et al. The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of Escherichia coli pathotypes. Mol. Microbiol. 2006, 61, 1556–1568. [Google Scholar] [CrossRef]
- Bertin, Y.; Girardeau, J.P.; Darfeuille-Michaud, A.; Contrepois, M. Characterization of 20K fimbria, a new adhesin of septicemic and diarrhea-associated Escherichia coli strains, that belongs to a family of adhesins with N-acetyl d-glucosamine recognition. Infect. Immun. 1996, 64, 332–342. [Google Scholar]
- Bertin, Y.; Martin, C.; Oswald, E.; Girardeau, J.P. Rapid and specific detection of F17-related pilin and adhesin genes in diarrheic and septicemic Escherichia coli strains by multiplex PCR. J. Clin. Microbiol. 1996, 34, 2921–2928. [Google Scholar]
- El Mazouari, K.; Oswald, E.; Hernalsteens, J.-P.; Lintermans, P.; de Greve, H. F-17-like fimbriae from an invasive Escherichia coli strain producing cytotoxic necrotizing factor type-2 toxin. Infect. Immun. 1994, 62, 2633–2638. [Google Scholar]
- Cid, D.; Sanz, R.; Marín, I.; de Greve, H.; Ruis-Santa-Quiteria, J.A.; Amils, R.; de la Fuente, R. Characterization of nonenterotoxigenic Escherichia coli strains producing F17 fimbriae isolated from diarrheic lambs and goat kids. J. Clin. Microbiol. 1999, 37, 1370–1375. [Google Scholar]
- Buts, L.; Wellens, A.; van Molle, I.; Wyns, L.; Loris, R.; Lahmann, M.; Oscarson, S.; de Greve, H.; Bouckaert, J. Impact of natural variation in bacterial F17G adhesins on crystallization behaviour. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 1149–1159. [Google Scholar] [CrossRef]
- Merckel, M.C.; Tanskanen, J.; Edelman, S.; Westerlund-Wikstrom, B.; Korhonen, T.K.; Goldman, A. The structural basis of receptor-binding by Escherichia coli associated with diarrhea and septicemia. J. Mol. Biol. 2003, 331, 897–905. [Google Scholar] [CrossRef]
- Buts, L.; Bouckaert, J.; de Genst, E.; Loris, R.; Oscarson, S.; Lahmann, M.; Messens, J.; Brosens, E.; Wyns, L.; de Greve, H. The fimbrial adhesin F17-G of enterotoxigenic Escherichia coli has an immunoglobulin-like lectin domain that binds N-acetylglucosamine. Mol. Microbiol. 2003, 49, 705–715. [Google Scholar]
- Imberechts, H.; Wild, P.; Charlier, G.; DeGreve, H.; Lintermans, P.; Pohl, P. Characterization of F18 fimbrial genes fedE and fedF involved in adhesion and length of enterotoxemic Escherichia coli strain 107/86. Microb. Pathog. 1996, 21, 183–192. [Google Scholar] [CrossRef]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.E.; Alvarez, R.; Bryan, M.C.; Fazio, F.; Calarese, D.; Stevens, J.; et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038. [Google Scholar] [CrossRef]
- De Boer, A.R.; Hokke, C.H.; Deelder, A.M.; Wuhrer, M. General Microarray Technique for Immobilization and Screening of Natural Glycans. Anal. Chem. 2007, 79, 8107–8113. [Google Scholar] [CrossRef]
- Lonardi, E.; Balog, C.I.; Deelder, A.M.; Wuhrer, M. Natural glycan microarrays. Exp. Rev. Proteomics. 2010, 7, 761–774. [Google Scholar] [CrossRef]
- Lonardi, E.; Deelder, A.M.; Wuhrer, M.; Balog, C.I. Microarray technology using glycans extracted from natural sources for serum antibody fluorescent detection. Methods Mol. Biol. 2012, 808, 285–302. [Google Scholar] [CrossRef]
- De Kerpel, M.; van Molle, I.; Brys, L.; Wyns, L.; de Greve, H.; Bouckaert, J. N-terminal truncation enables crystallization of the receptor-binding domain of the FedF bacterial adhesin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 1278–1282. [Google Scholar] [CrossRef]
- Moonens, K.; Bouckaert, J.; Coddens, A.; Tran, T.; Panjikar, S.; de Kerpel, M.; Cox, E.; Remaut, H.; de Greve, H. Structural insight in histo-blood group binding by the F18 fimbrial adhesin FedF. Mol. Microbiol. 2012, 86, 82–95. [Google Scholar] [CrossRef]
- Wellens, A.; Garofalo, C.; Nguyen, H.; van Gerven, N.; Slattegard, R.; Hernalsteens, J.P.; Wyns, L.; Oscarson, S.; de Greve, H.; Hultgren, S.; et al. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS One 2008, 3, e2040. [Google Scholar] [CrossRef]
- CFG Functional Glycomics Gateway, Glycan Array Data. Available online: http://www.functionalglycomics.org/glycomics/publicdata/primaryscreen.jsp/ (accessed on 10 September 2010).
- Cirla, A.; McHale, A.R.; Mann, J. Synthesis of analogues of calicheamicin and neocarzinostatin chromophore. Tetrahedron 2004, 60, 4019–4029. [Google Scholar] [CrossRef]
- Wang, H.; She, J.; Zhang, L.H.; Ye, X.S. Silver(I) oxide mediated selective monoprotection of diols in pyranosides. J. Org. Chem. 2004, 69, 5774–5777. [Google Scholar] [CrossRef]
- Kohata, K.; Abbas, S.A.; Matta, K.L. Synthetic mucin fragments: Methyl 3-O-(2-acetamido-2-deoxy-β-d-galactopyranosyl-β-d-3-O-(2-acetamido-2-deoxy-3-O-β-d-galactopyranosyl-β-d-glucopyranosyl)-β-d-galactoyranoside. pyranosyl)-β-d-galactopyranoside. Carbohydr. Res. 1984, 132, 127–135. [Google Scholar] [CrossRef]
- Garegg, P.J.; Oscarson, S. Synthesis of 6- and 6'-deoxy derivatives of methyl 4-O-a-galactopyranosyl-ß-galactopyranoside for studies of inhibition of pyelonephritogenic fimbriated E. coli adhesion to urinary epithelium-cell surfaces. Carbohydr. Res. 1985, 137, 270–275. [Google Scholar] [CrossRef]
- Valdor, J.F.; Mackie, W. Synthesis of a trisaccharide repeating unit related to arabinogalactan-protein (AGP) polysaccharides. J. Carbohydr. Chem. 1997, 16, 429–440. [Google Scholar] [CrossRef]
- Kajihara, Y.; Kodama, H.; Endo, T.; Hashimoto, H. Novel features of acceptor recognition by ß-(1-4)-galactosyltransferase. Carbohydr. Res. 1998, 306, 361–378. [Google Scholar] [CrossRef]
- Abbas, S.A.; Kohata, K.; Matta, K.L. Synthetic mucin fragments. Methyl 6-O-(2-acetamido-2-deoxy-ß-glucopyranosyl)-ß-d-galactopyranoside, methyl 3,4-di-O-(2-acetamido-2-deoxy-ß-d-glucopyranosyl)-ß-d-galactopyranoside, and methyl O-(2-acetamido-2-deoxy-ß-d-glucopyranosyl)-(1–3)-O-ß-d-galactopyranosyl-(1–3)-O-(2-acetamido-2-deoxy-ß-d-glucopyranosyl)-(1–3)-ß-d-galactopyranoside. Carbohydr. Res. 1987, 161, 39–47. [Google Scholar] [CrossRef]
- Wuhrer, M.; Koeleman, C.A.; Hokke, C.H.; Deelder, A.M. Mass spectrometry of proton adducts of fucosylated N-glycans: Fucose transfer between antennae gives rise to misleading fragments. Rapid Commun. Mass Spectrom. 2006, 20, 1747–1754. [Google Scholar] [CrossRef]
- Taganna, J.; de Boer, A.R.; Wuhrer, M.; Bouckaert, J. Glycosylation changes as important factors for the susceptibility to urinary tract infection. Biochem. Soc. Trans. 2011, 39, 349–354. [Google Scholar] [CrossRef]
- Dauter, M.; Dauter, Z. Phase determination using halide ions. Methods Mol. Biol. 2007, 364, 149–158. [Google Scholar]
- Coddens, A.; Diswall, M.; Angstrom, J.; Breimer, M.E.; Goddeeris, B.; Cox, E.; Teneberg, S. Recognition of blood group ABH type 1 determinants by the FedF adhesin of F18-fimbriated Escherichia coli. J. Biol. Chem. 2009, 284, 9713–9726. [Google Scholar] [CrossRef]
- Meijerink, E.; Neuenschwander, S.; Fries, R.; Dinter, A.; Bertschinger, H.U.; Stranzinger, G.; Vogeli, P. A DNA polymorphism influencing alpha(1,2)fucosyltransferase activity of the pig FUT1 enzyme determines susceptibility of small intestinal epithelium to Escherichia coli F18 adhesion. Immunogenetics 2000, 52, 129–136. [Google Scholar] [CrossRef]
- Hung, C.-S.; Bouckaert, J.; Hung, D.L.; Pinkner, J.; Winberg, C.; Defusco, A.; Auguste, C.G.; Strouse, R.; Langermann, S.; Waksman, G.; et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 2002, 44, 903–915. [Google Scholar] [CrossRef]
- Mouricout, M.A.; Julien, R.A. Pilus-mediated binding of bovine enterotoxigenic Escherichia coli to calf small intestinal mucins. Infect. Immun. 1987, 55, 1216–1223. [Google Scholar]
- Stgeme, J.W.; Cutter, D. Evidence That Surface Fibrils Expressed by Haemophilus-Influenzae Type-B Promote Attachment to Human Epithelial-Cells. Mol. Microbiol. 1995, 15, 77–85. [Google Scholar] [CrossRef]
- Nuccio, S.P.; Baumler, A.J. Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 2007, 71, 551–575. [Google Scholar] [CrossRef]
- Berenson, C.S.; Sayles, K.B.; Huang, J.; Reinhold, V.N.; Garlipp, M.A.; Yohe, H.C. Nontypeable Haemophilus influenzae-binding gangliosides of human respiratory (HEp-2) cells have a requisite lacto/neolacto core structure. FEMS Immunol. Med. Microbiol. 2005, 45, 171–182. [Google Scholar] [CrossRef]
- Mouricout, M.; Milhavet, M.; Durie, C.; Grange, P. Characterization of glycoprotein glycan receptors for Escherichia coli F17 fimbrial lectin. Microb. Pathog. 1995, 18, 297–306. [Google Scholar] [CrossRef]
- Vercoutter-Edouart, A.S.; Slomianny, M.C.; Dekeyzer-Beseme, O.; Haeuw, J.F.; Michalski, J.C. Glycoproteomics and glycomics investigation of membrane N-glycosylproteins from human colon carcinoma cells. Proteomics 2008, 8, 3236–3256. [Google Scholar] [CrossRef]
- Salo, H.; Aitio, O.; Ilves, K.; Bencomo, E.; Toivonen, S.; Penttila, L.; Niemela, R.; Salminen, H.; Grabenhorst, E.; Renkonen, R.; et al. Several polylactosamine-modifying glycosyltransferases also use internal GalNAcbeta1-4GlcNAc units of synthetic saccharides as acceptors. Glycobiology 2002, 12, 217–228. [Google Scholar] [CrossRef]
- Dam, T.K.; Brewer, C.F. Lectins as pattern recognition molecules: The effects of epitope density in innate immunity. Glycobiology 2010, 20, 270–279. [Google Scholar] [CrossRef]
- Robbe, C.; Capon, C.; Maes, E.; Rousset, M.; Zweibaum, A.; Zanetta, J.P.; Michalski, J.C. Evidence of regio-specific glycosylation in human intestinal mucins: Presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 2003, 278, 46337–46348. [Google Scholar]
- Robbe, C.; Capon, C.; Coddeville, B.; Michalski, J.C. Diagnostic ions for the rapid analysis by nano-electrospray ionization quadrupole time-of-flight mass spectrometry of O-glycans from human mucins. Rapid Commun. Mass Spectrom. 2004, 18, 412–420. [Google Scholar] [CrossRef]
- Teneberg, S.; Willemsen, P.T.; de Graaf, F.K.; Stenhagen, G.; Pimlott, W.; Jovall, P.A.; Angstrom, J.; Karlsson, K.A. Characterization of gangliosides of epithelial cells of calf small intestine, with special reference to receptor-active sequences for enteropathogenic Escherichia coli K99. J. Biochem. 1994, 116, 560–574. [Google Scholar]
- Bouckaert, J.; Hamelryck, T.; Wyns, L.; Loris, R. Novel structures of plant lectins and their complexes with carbohydrates. Curr. Opin. Struct. Biol. 1999, 9, 572–577. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lonardi, E.; Moonens, K.; Buts, L.; De Boer, A.R.; Olsson, J.D.M.; Weiss, M.S.; Fabre, E.; Guérardel, Y.; Deelder, A.M.; Oscarson, S.; et al. Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli. Biology 2013, 2, 894-917. https://doi.org/10.3390/biology2030894
Lonardi E, Moonens K, Buts L, De Boer AR, Olsson JDM, Weiss MS, Fabre E, Guérardel Y, Deelder AM, Oscarson S, et al. Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli. Biology. 2013; 2(3):894-917. https://doi.org/10.3390/biology2030894
Chicago/Turabian StyleLonardi, Emanuela, Kristof Moonens, Lieven Buts, Arjen R. De Boer, Johan D. M. Olsson, Manfred S. Weiss, Emeline Fabre, Yann Guérardel, André M. Deelder, Stefan Oscarson, and et al. 2013. "Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli" Biology 2, no. 3: 894-917. https://doi.org/10.3390/biology2030894
APA StyleLonardi, E., Moonens, K., Buts, L., De Boer, A. R., Olsson, J. D. M., Weiss, M. S., Fabre, E., Guérardel, Y., Deelder, A. M., Oscarson, S., Wuhrer, M., & Bouckaert, J. (2013). Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli. Biology, 2(3), 894-917. https://doi.org/10.3390/biology2030894







