Mitotic Spindle Assembly in Land Plants: Molecules and Mechanisms
Abstract
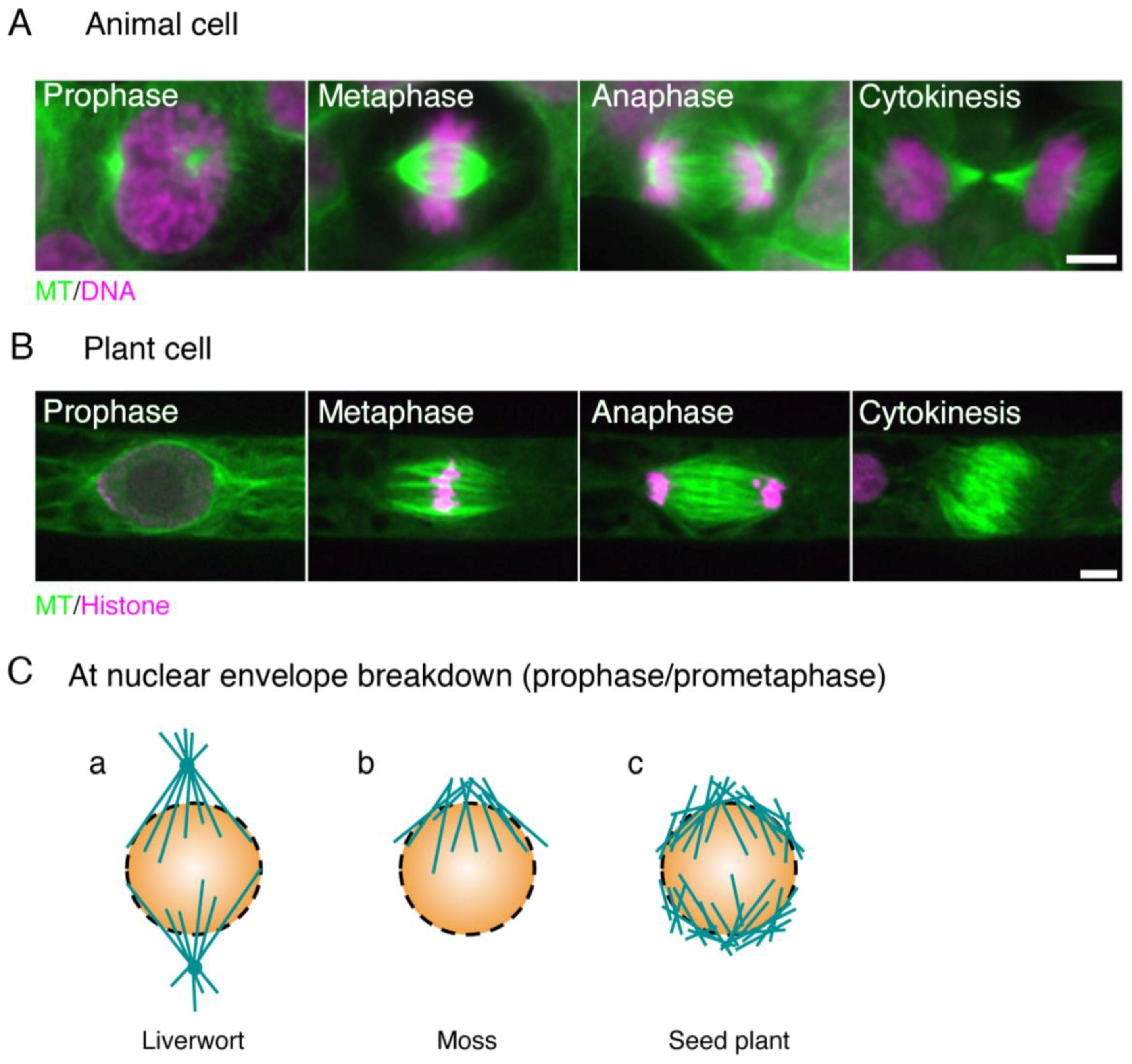
:1. Microscopic Overview of the Spindle Assembly
1.1. Mitotic Spindle Assembly in Animals
1.2. Mitotic Spindle Assembly in Seed Plants
1.3. Mitotic Spindle Assembly in Bryophytes
2. Conservation of Spindle Assembly Factors
- The amino acid sequences of the Homo sapiens proteins were retrieved from the NCBI database. When multiple isoforms were identified, only one randomly selected isoform was used.
- Drosophila melanogaster (fruit fly) and Schizosaccharomyces. pombe (fission yeast) homologues were sought in the NCBI ‘Homologene’ or ‘Gene’ search. When clear homologues were not identified, the BLAST search was performed.
- Homologous genes of Arabidopsis thaliana and Physcomitrella. patens were sought using BLAST (query: human or yeast protein).
- If no clear homologues could be identified, the databases for individual species were searched (PomBase, fly base, PHYSCObase, or TAIR). For the query, human (or, in some instances, fly) gene names or keywords (e.g., ‘centromere’, ‘kinetochore’, or ‘CENP’) were used.
- If homologous genes were still not identified, the name was searched using Google Scholar and PubMed.
- The sequences of plant Dsn1/Nnf1/Spc24/Ska3 and Msd1 were provided by Dr. Geert Kops (Utrecht University, The Netherlands) and Dr. Takashi Hashimoto (Nara Institute of Science and Technology, Japan), respectively.
2.1. Centrosome Proteins
2.2. Dynein Complex and Its Localisation Factors
2.3. Kinetochore Components
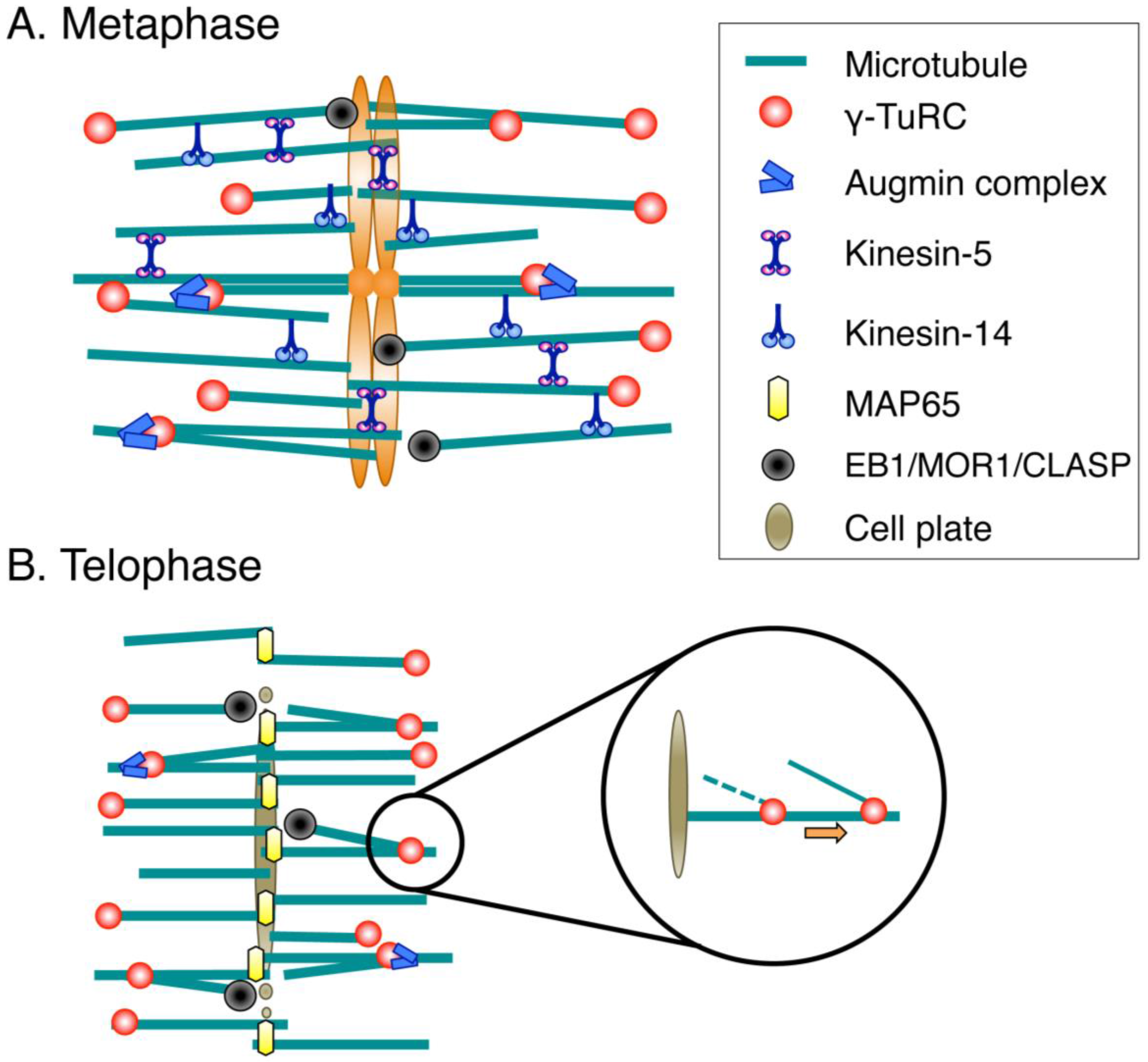
3. Molecular Mechanisms of Spindle Assembly in Land Plants
3.1. Spindle Assembly
3.2. Phragmoplast Assembly
4. Conclusions and Future Perspectives on Spindle Research in Plants
Acknowledgments
Conflicts of Interest
References
- Inoue, S.; Sato, H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 1967, 50, S259–S292. [Google Scholar] [CrossRef]
- Karp, G. Cell and Molecular Biology: Concepts and Experiments, 6th ed.; John Wiley: Hoboken, NJ, USA, 2010. [Google Scholar]
- Conduit, P.T.; Richens, J.H.; Wainman, A.; Holder, J.; Vicente, C.C.; Pratt, M.B.; Dix, C.I.; Novak, Z.A.; Dobbie, I.M.; Schermelleh, L.; et al. A molecular mechanism of mitotic centrosome assembly in drosophila. eLife 2014, 3, e03399. [Google Scholar] [CrossRef] [PubMed]
- Duncan, T.; Wakefield, J.G. 50 ways to build a spindle: The complexity of microtubule generation during mitosis. Chromosome Res. 2011, 19, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Reber, S.; Hyman, A.A. Emergent properties of the metaphase spindle. Cold Spring Harb. Perspect. Biol. 2015, 7, a015784. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Heald, R. Mechanisms of mitotic spindle assembly and function. Int. Rew. Cytol. 2008, 265, 111–158. [Google Scholar]
- Goshima, G.; Kimura, A. New look inside the spindle: Microtubule-Dependent microtubule generation within the spindle. Curr. Opin. Cell Biol. 2010, 22, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Hayward, D.; Metz, J.; Pellacani, C.; Wakefield, J.G. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell 2014, 28, 81–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walczak, C.E.; Vernos, I.; Mitchison, T.J.; Karsenti, E.; Heald, R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998, 8, 903–913. [Google Scholar] [CrossRef]
- Hatsumi, M.; Endow, S.A. Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J. Cell Sci. 1992, 101, 547–559. [Google Scholar] [PubMed]
- Wakefield, J.G.; Bonaccorsi, S.; Gatti, M. The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 2001, 153, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Scholey, J.M. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 2010, 26, 21–57. [Google Scholar] [CrossRef] [PubMed]
- Uehara, R.; Goshima, G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol. 2010, 191, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Glotzer, M. The 3ms of central spindle assembly: Microtubules, motors and maps. Nat. Rev. Mol. Cell Biol. 2009, 10, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.C.; Lemmon, B.E. Dividing without centrioles: Innovative plant microtubule organizing centres organize mitotic spindles in bryophytes, the earliest extant lineages of land plants. AoB Plants 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, H.; Holtmannspotter, M.; Borchers, A.; O’Donoghue, M.T.; Zachgo, S. Microtubule dynamics of the centrosome-like polar organizers from the basal land plant marchantia polymorpha. New Phytol. 2016, 209, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, Y.; Miki, T.; Fujioka, R.; Uehara, R.; Tomioka, A.; Obuse, C.; Kubo, M.; Hiwatashi, Y.; Goshima, G. An inducible rna interference system in physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell 2012, 24, 1478–1493. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, C.; Chan, J. Not so divided: The common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 2006, 7, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.; Calder, G.; Fox, S.; Lloyd, C. Localization of the microtubule end binding protein eb1 reveals alternative pathways of spindle development in arabidopsis suspension cells. Plant Cell 2005, 17, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Baskin, T.I.; Cande, W.Z. The structure and function of the mitotic spindle in flowering plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1990, 41, 277–315. [Google Scholar] [CrossRef]
- Smirnova, E.A.; Bajer, A.S. Spindle poles in higher plant mitosis. Cell Motil Cytoskelet. 1992, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- De Mey, J.; Lambert, A.M.; Bajer, A.S.; Moeremans, M.; De Brabander, M. Visualization of microtubules in interphase and mitotic plant cells of haemanthus endosperm with the immuno-gold staining method. Proc. Natl. Acad. Sci. USA 1982, 79, 1898–1902. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.A.; Bajer, A.S. Microtubule converging centers and reorganization of the interphase cytoskeleton and the mitotic spindle in higher plant haemanthus. Cell Motil Cytoskelet. 1994, 27, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.A.; Bajer, A.S. Early stages of spindle formation and independence of chromosome and microtubule cycles in haemanthus endosperm. Cell Motil Cytoskelet. 1998, 40, 22–37. [Google Scholar] [CrossRef]
- Falconer, M.M.; Donaldson, G.; Seagull, R.W. Mtocs in higher-plant cells—An immunofluorescent study of microtubule assembly sites following depolymerization by apm. Protoplasma 1988, 144, 46–55. [Google Scholar] [CrossRef]
- Euteneuer, U.; Jackson, W.T.; McIntosh, J.R. Polarity of spindle microtubules in haemanthus endosperm. J. Cell Biol. 1982, 94, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Bajer, A.S.; Mole-Bajer, J. Reorganization of microtubules in endosperm cells and cell fragments of the higher plant haemanthus in vivo. J. Cell Biol. 1986, 102, 263–281. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.M.; Bajer, A.S. Dynamics of spindle fibers and microtubules during anaphase and phragmoplast formation. Chromosoma 1972, 39, 101–144. [Google Scholar] [CrossRef]
- Liu, B.; Ho, C.M.; Lee, Y.R. Microtubule reorganization during mitosis and cytokinesis: Lessons learned from developing microgametophytes in arabidopsis thaliana. Front. Plant Sci. 2011. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, J.C.; Cyr, R. The kinesin atk5 functions in early spindle assembly in arabidopsis. Plant Cell. 2007, 19, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Sano, T.; Kutsuna, N.; Kumagai-Sano, F.; Hasezawa, S. Contribution of anaphase b to chromosome separation in higher plant cells estimated by image processing. Plant Cell Physiol. 2007, 48, 1509–1513. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, J.C.; Cyr, R. Mitotic spindle organization by the preprophase band. Mol. Plant 2008, 1, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Wright, A.J.; Smith, L.G. Division plane control in plants: New players in the band. Trends Cell Biol. 2009, 19, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, C.G.; Humphries, J.A.; Smith, L.G. Determination of symmetric and asymmetric division planes in plant cells. Annu. Rev. Plant Biol. 2011, 62, 387–409. [Google Scholar] [CrossRef] [PubMed]
- Fowke, L.C.; Pickett-Heaps, J.D. Electron microscope study of vegetative cell division in two species of marchantia. Can. J. Bot. 1978, 56, 467–475. [Google Scholar] [CrossRef]
- Brown, R.C.; Lemmon, B.E. Polar organizers mark division axis prior to preprophase band formation in mitosis of the hepaticreboulia hemisphaerica (bryophyta). Protoplasma 1990, 156, 74–81. [Google Scholar] [CrossRef]
- Shimamura, M.; Brown, R.C.; Lemmon, B.E.; Akashi, T.; Mizuno, K.; Nishihara, N.; Tomizawa, K.; Yoshimoto, K.; Deguchi, H.; Hosoya, H.; et al. Gamma-Tubulin in basal land plants: Characterization, localization, and implication in the evolution of acentriolar microtubule organizing centers. Plant Cell 2004, 16, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.C.; Lemmon, B.E.; Horio, T. Gamma-Tubulin localization changes from discrete polar organizers to anastral spindles and phragmoplasts in mitosis of marchantia polymorpha l. Protoplasma 2004, 224, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.C.; Lemmon, B.E. Preprophase microtubule systems and development of the mitotic spindle in hornworts (bryophyta). Protoplasma 1988, 143, 11–21. [Google Scholar] [CrossRef]
- Goshima, G.; Wollman, R.; Goodwin, N.; Zhang, J.M.; Scholey, J.M.; Vale, R.D.; Stuurman, N. Genes required for mitotic spindle assembly in drosophila s2 cells. Science 2007, 316, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Sonnichsen, B.; Koski, L.B.; Walsh, A.; Marschall, P.; Neumann, B.; Brehm, M.; Alleaume, A.M.; Artelt, J.; Bettencourt, P.; Cassin, E.; et al. Full-Genome rnai profiling of early embryogenesis in caenorhabditis elegans. Nature 2005, 434, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, J.R.; Toyoda, Y.; Hegemann, B.; Poser, I.; Heriche, J.K.; Sykora, M.M.; Augsburg, M.; Hudecz, O.; Buschhorn, B.A.; Bulkescher, J.; et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010, 328, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, M.A.; Geiser, J.R. Genetic analysis of the mitotic spindle. Annu. Rev. Genet. 1996, 30, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Genome, I. Analysis of the genome sequence of the flowering plant arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T. Microtubule organization and microtubule-associated proteins in plant cells. Int. Rev. Cell Mol. Biol. 2014, 312, 1–52. [Google Scholar] [PubMed]
- Gardiner, J. The evolution and diversification of plant microtubule-associated proteins. Plant J. 2013, 75, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Demidov, D.; Van Damme, D.; Geelen, D.; Blattner, F.R.; Houben, A. Identification and dynamics of two classes of aurora-like kinases in arabidopsis and other plants. Plant Cell 2005, 17, 836–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Carcer, G.; Manning, G.; Malumbres, M. From plk1 to plk5: Functional evolution of polo-like kinases. Cell Cycle 2011, 10, 2255–2262. [Google Scholar] [CrossRef] [PubMed]
- Moorhead, G.B.; De Wever, V.; Templeton, G.; Kerk, D. Evolution of protein phosphatases in plants and animals. Biochem. J. 2009, 417, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Labandera, A.M.; Vahab, A.R.; Chaudhuri, S.; Kerk, D.; Moorhead, G.B. The mitotic pp2a regulator ensa/arpp-19 is remarkably conserved across plants and most eukaryotes. Biochem. Biophys. Res. Commun. 2015, 458, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Santos, Z.; Machado, P.; Branco, P.; Tavares-Cadete, F.; Rodrigues-Martins, A.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 2010, 123, 1414–1426. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Dixit, R. Functions of the arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 2012, 249, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Vanstraelen, M.; Inze, D.; Geelen, D. Mitosis-specific kinesins in arabidopsis. Trends Plant Sci. 2006, 11, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Wickstead, B.; Gull, K. Dyneins across eukaryotes: A comparative genomic analysis. Traffic 2007, 8, 1708–1721. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Plant actin-related proteins. Trends Plant Sci. 2004, 9, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Vos, J.W.; Pieuchot, L.; Evrard, J.L.; Janski, N.; Bergdoll, M.; de Ronde, D.; Perez, L.H.; Sardon, T.; Vernos, I.; Schmit, A.C. The plant tpx2 protein regulates prospindle assembly before nuclear envelope breakdown. Plant Cell 2008, 20, 2783–2797. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Joshi, H.C.; Wilson, T.J.; Silflow, C.D.; Palevitz, B.A.; Snustad, D.P. Gamma-tubulin in arabidopsis: Gene sequence, immunoblot, and immunofluorescence studies. Plant Cell 1994, 6, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Hotta, T.; Lee, Y.R.; Horio, T.; Liu, B. The {gamma}-tubulin complex protein gcp4 is required for organizing functional microtubule arrays in arabidopsis thaliana. Plant Cell 2010, 22, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.J.; Lee, Y.R.; Liu, B. The wd40 repeat protein nedd1 functions in microtubule organization during cell division in arabidopsis thaliana. Plant Cell 2009, 21, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Batzenschlager, M.; Lermontova, I.; Schubert, V.; Fuchs, J.; Berr, A.; Koini, M.A.; Houlne, G.; Herzog, E.; Rutten, T.; Alioua, A.; et al. Arabidopsis mzt1 homologs gip1 and gip2 are essential for centromere architecture. Proc. Natl. Acad. Sci. USA 2015, 112, 8656–8660. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Yagi, N.; Kato, T.; Fujita, S.; Kawashima, N.; Ehrhardt, D.W.; Hashimoto, T. Arabidopsis gcp3-interacting protein 1/mozart 1 is an integral component of the gamma-tubulin-containing microtubule nucleating complex. Plant J. 2012, 71, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Janski, N.; Masoud, K.; Batzenschlager, M.; Herzog, E.; Evrard, J.L.; Houlne, G.; Bourge, M.; Chaboute, M.E.; Schmit, A.C. The gcp3-interacting proteins gip1 and gip2 are required for gamma-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell 2012, 24, 1171–1187. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C.; Neuner, A.; Schiebel, E. Targeting of gamma-tubulin complexes to microtubule organizing centers: Conservation and divergence. Trends Cell Biol. 2015, 25, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Hotta, T.; Kong, Z.; Ho, C.M.; Zeng, C.J.; Horio, T.; Fong, S.; Vuong, T.; Lee, Y.R.; Liu, B. Characterization of the arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell 2012, 24, 1494–1509. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, S.; Zheng, Y. Characterization of a drosophila centrosome protein cp309 that shares homology with kendrin and cg-nap. Mol. Biol. Cell 2004, 15, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, E.; Wasteneys, G.O. Mor1, the arabidopsis thaliana homologue of xenopus map215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J. Cell Sci. 2008, 121, 4114–4123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Hua, S.; Mohan, R.; Grigoriev, I.; Yau, K.W.; Liu, Q.; Katrukha, E.A.; Altelaar, A.F.; Heck, A.J.; Hoogenraad, C.C.; et al. Microtubule minus-end stabilization by polymerization-driven camsap deposition. Dev. Cell 2014, 28, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Nagasaki-Takeuchi, N.; Kato, T.; Fujiwara, M.; Sonobe, S.; Fukao, Y.; Hashimoto, T. Purification and characterization of novel microtubule-associated proteins from arabidopsis cell suspension cultures. Plant Physiol. 2013, 163, 1804–1816. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. Condensins: Organizing and segregating the genome. Curr. Biol. 2005, 15, R265–R275. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, N.U. Mutations in arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 2003, 130, 3283–3295. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.; Yonemura, M.; Matsunaga, S.; Nakagawa, T.; Uchiyama, S.; Fukui, K. Characterization and dynamic analysis of arabidopsis condensin subunits, atcap-h and atcap-h2. Planta 2005, 222, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, N.U.; Rusyniak, S.; Hasenkampf, C.A.; Riggs, C.D. Disruption of the arabidopsis smc4 gene, atcap-c, compromises gametogenesis and embryogenesis. Planta 2006, 223, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.M.; Lister, C.; Page, T.; Fransz, P.; Findlay, K.; Jones, G.H.; Dickinson, H.G.; Dean, C. The dif1 gene of arabidopsis is required for meioticchromosome segregation and belongs to the rec8/rad21cohesin gene family. Plant J. 1999, 19, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Schubert, V.; Weissleder, A.; Ali, H.; Fuchs, J.; Lermontova, I.; Meister, A.; Schubert, I. Cohesin gene defects may impair sister chromatid alignment and genome stability in arabidopsis thaliana. Chromosoma 2009, 118, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, J.; Ravi, M.; Andreuzza, S.; Panoli, A.P.; Marimuthu, M.P.; Siddiqi, I. The plant adherin atscc2 is required for embryogenesis and sister-chromatid cohesion during meiosis in arabidopsis. Plant J. 2009, 59, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Andreuzza, S.; Panoli, A.P.; Siddiqi, I. Atctf7 is required for establishment of sister chromatid cohesion and association of cohesin with chromatin during meiosis in arabidopsis. BMC Plant Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Ladurner, R.; Kreidl, E.; Ivanov, M.P.; Ekker, H.; Idarraga-Amado, M.H.; Busslinger, G.A.; Wutz, G.; Cisneros, D.A.; Peters, J.M. Sororin actively maintains sister chromatid cohesion. EMBO J. 2016, 35, 635–653. [Google Scholar] [CrossRef] [PubMed]
- Zamariola, L.; De Storme, N.; Tiang, C.L.; Armstrong, S.J.; Franklin, F.C.; Geelen, D. Sgo1 but not sgo2 is required for maintenance of centromere cohesion in arabidopsis thaliana meiosis. Plant Reprod. 2013, 26, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Kirioukhova, O.; Johnston, A.J.; Kleen, D.; Kagi, C.; Baskar, R.; Moore, J.M.; Baumlein, H.; Gross-Hardt, R.; Grossniklaus, U. Female gametophytic cell specification and seed development require the function of the putative arabidopsis incenp ortholog wyrd. Development 2011, 138, 3409–3420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lermontova, I.; Kuhlmann, M.; Friedel, S.; Rutten, T.; Heckmann, S.; Sandmann, M.; Demidov, D.; Schubert, V.; Schubert, I. Arabidopsis kinetochore null2 is an upstream component for centromeric histone h3 variant cenh3 deposition at centromeres. Plant Cell 2013, 25, 3389–3404. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Masuelli, R.; Tyagi, A.P.; Comai, L.; Henikoff, S. Centromeric localization and adaptive evolution of an arabidopsis histone h3 variant. Plant Cell 2002, 14, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Shibata, F.; Sato, H.; Murata, M. Characterization of a cenp-c homolog in arabidopsis thaliana. Genes Genet. Syst. 2004, 79, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Dangel, N.J.; Knoll, A.; Puchta, H. Mhf1 plays fanconi anaemia complementation group M protein (fancm)-dependent and fancm-independent roles in DNA repair and homologous recombination in plants. Plant J. 2014, 78, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Girard, C.; Crismani, W.; Froger, N.; Mazel, J.; Lemhemdi, A.; Horlow, C.; Mercier, R. Fancm-associated proteins mhf1 and mhf2, but not the other fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 2014, 42, 9087–9095. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Shibata, F.; Murata, M. Characterization of a mis12 homologue in arabidopsis thaliana. Chromosome Res. 2005, 13, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Tromer, E.; Snel, B.; Kops, G.J. Widespread recurrent patterns of rapid repeat evolution in the kinetochore scaffold knl1. Genome Biol. Evol. 2015, 7, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, M.C.; Paganelli, L.; Lecomte, P.; Deslandes, L.; Quentin, M.; Pecrix, Y.; Le Bris, M.; Marfaing, N.; Abad, P.; Favery, B. Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS ONE 2009, 4, e6757. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tang, D.; Luo, Q.; Jin, Y.; Shen, Y.; Wang, K.; Cheng, Z. Brk1, a bub1-related kinase, is essential for generating proper tension between homologous kinetochores at metaphase i of rice meiosis. Plant Cell 2012, 24, 4961–4973. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, E.A.; Romeiro, N.C.; Ribeiro Eda, S.; Santa-Catarina, C.; Oliveira, A.E.; Silveira, V.; de Souza Filho, G.A.; Venancio, T.M.; Cruz, M.A. Structural and functional characterization of the protein kinase mps1 in arabidopsis thaliana. PLoS ONE 2012, 7, e45707. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Rose, A.; Muthuswamy, S.; Jeong, S.Y.; Venkatakrishnan, S.; Zhao, Q.; Meier, I. Nuclear pore anchor, the arabidopsis homolog of tpr/mlp1/mlp2/megator, is involved in mrna export and sumo homeostasis and affects diverse aspects of plant development. Plant Cell 2007, 19, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A.; Williams, B.C.; Li, Z.; Etemad-Moghadam, B.; Dawe, R.K.; Goldberg, M.L. Conservation of the centromere/kinetochore protein zw10. J. Cell Biol. 1997, 138, 1289–1301. [Google Scholar] [CrossRef] [PubMed]
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Drykova, D.; Cenklova, V.; Sulimenko, V.; Volc, J.; Draber, P.; Binarova, P. Plant gamma-tubulin interacts with alphabeta-tubulin dimers and forms membrane-associated complexes. Plant Cell 2003, 15, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Sonobe, S.; Baskin, T.I.; Hyodo, S.; Hasezawa, S.; Nagata, T.; Horio, T.; Hasebe, M. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 2005, 7, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, J.; Nacry, P.; Christodoulidou, A.; Drevensek, S.; Camilleri, C.; Amiour, N.; Parcy, F.; Pastuglia, M.; Bouchez, D. Arabidopsis tonneau1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 2008, 20, 2146–2159. [Google Scholar] [CrossRef] [PubMed]
- Hodges, M.E.; Wickstead, B.; Gull, K.; Langdale, J.A. The evolution of land plant cilia. New Phytol. 2012, 195, 526–540. [Google Scholar] [CrossRef] [PubMed]
- Kardon, J.R.; Vale, R.D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009, 10, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, C.J.; Morris, N.R.; Meagher, R.B.; Dawe, R.K. Dyneins have run their course in plant lineage. Traffic 2001, 2, 362–363. [Google Scholar] [CrossRef] [PubMed]
- Rapali, P.; Szenes, A.; Radnai, L.; Bakos, A.; Pal, G.; Nyitray, L. Dynll/lc8: A light chain subunit of the dynein motor complex and beyond. FEBS J. 2011, 278, 2980–2996. [Google Scholar] [CrossRef] [PubMed]
- Endow, S.A. Determinants of molecular motor directionality. Nat. Cell Biol. 1999, 1, E163–E167. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, E.; Yamada, M.; Vale, R.D.; Goshima, G. Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants. Nat. Plants 2015, 1. [Google Scholar] [CrossRef] [PubMed]
- Furuta, K.; Furuta, A.; Toyoshima, Y.Y.; Amino, M.; Oiwa, K.; Kojima, H. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc. Natl. Acad. Sci. USA 2013, 110, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Nedelec, F.; Vale, R.D. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005, 171, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Hyams, J.S.; Campbell, C.J. Widespread absence of outer dynein arms in the spermatozoids of lower plants. Cell Biol. Int. Rep. 1985, 9, 841–848. [Google Scholar] [CrossRef]
- Komaki, S.; Schnittger, A. The spindle checkpoint in plants-a green variation over a conserved theme? Curr. Opin. Plant Biol. 2016, 34, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Przewloka, M.R.; Zhang, W.; Costa, P.; Archambault, V.; D’Avino, P.P.; Lilley, K.S.; Laue, E.D.; McAinsh, A.D.; Glover, D.M. Molecular analysis of core kinetochore composition and assembly in drosophila melanogaster. PLoS ONE 2007, 2, e478. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Nishina, M.; Goshima, G. Rnai screening identifies the armadillo repeat-containing kinesins responsible for microtubule-dependent nuclear positioning in physcomitrella patens. Plant Cell Physiol. 2015, 56, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Binarova, P.; Cenklova, V.; Prochazkova, J.; Doskocilova, A.; Volc, J.; Vrlik, M.; Bogre, L. Gamma-Tubulin is essential for acentrosomal microtubule nucleation and coordination of late mitotic events in arabidopsis. Plant Cell 2006, 18, 1199–1212. [Google Scholar] [CrossRef] [PubMed]
- Pastuglia, M.; Azimzadeh, J.; Goussot, M.; Camilleri, C.; Belcram, K.; Evrard, J.L.; Schmit, A.C.; Guerche, P.; Bouchez, D. Gamma-Tubulin is essential for microtubule organization and development in arabidopsis. Plant Cell 2006, 18, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Mayer, M.; Zhang, N.; Stuurman, N.; Vale, R.D. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 2008, 181, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Tian, J.; Wang, G.; Yu, Y.; Wang, C.; Ma, Y.; Zhang, X.; Xia, G.; Liu, B.; Kong, Z. Augmin triggers microtubule-dependent microtubule nucleation in interphase plant cells. Curr. Biol. 2014, 24, 2708–2713. [Google Scholar] [CrossRef] [PubMed]
- Bannigan, A.; Scheible, W.R.; Lukowitz, W.; Fagerstrom, C.; Wadsworth, P.; Somerville, C.; Baskin, T.I. A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 2007, 120, 2819–2827. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Naito, H.; Nishina, M.; Goshima, G. Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc. Natl. Acad. Sci. USA 2014, 111, E1053–E1061. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Marcus, A.; Li, W.; Hu, Y.; Calzada, J.P.; Grossniklaus, U.; Cyr, R.J.; Ma, H. The arabidopsis atk1 gene is required for spindle morphogenesis in male meiosis. Development 2002, 129, 2401–2409. [Google Scholar] [PubMed]
- Ambrose, J.C.; Li, W.; Marcus, A.; Ma, H.; Cyr, R. A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 2005, 16, 1584–1592. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.I.; Li, W.; Ma, H.; Cyr, R.J. A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol. Biol. Cell 2003, 14, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Goshima, G. Microcephaly protein asp focuses the minus ends of spindle microtubules at the pole and within the spindle. J. Cell Biol. 2015, 211, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, E.; Himmelspach, R.; Rashbrooke, M.C.; Whittington, A.T.; Gale, K.R.; Collings, D.A.; Wasteneys, G.O. Microtubule organization 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the arabidopsis root. Plant Physiol. 2006, 140, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Komaki, S.; Abe, T.; Coutuer, S.; Inze, D.; Russinova, E.; Hashimoto, T. Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in arabidopsis. J. Cell Sci. 2010, 123, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiato, H.; Khodjakov, A.; Rieder, C.L. Drosophila clasp is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 2005, 7, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, J.C.; Shoji, T.; Kotzer, A.M.; Pighin, J.A.; Wasteneys, G.O. The arabidopsis clasp gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 2007, 19, 2763–2775. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.C.; Rogers, S.L.; Sharp, D.J. Spindle microtubules in flux. J. Cell Sci. 2005, 118, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Dhonukshe, P.; Vischer, N.; Gadella, T.W., Jr. Contribution of microtubule growth polarity and flux to spindle assembly and functioning in plant cells. J. Cell Sci. 2006, 119, 3193–3205. [Google Scholar] [CrossRef] [PubMed]
- Otegui, M.S.; Verbrugghe, K.J.; Skop, A.R. Midbodies and phragmoplasts: Analogous structures involved in cytokinesis. Trends Cell Biol. 2005, 15, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Duellberg, C.; Fourniol, F.J.; Maurer, S.P.; Roostalu, J.; Surrey, T. End-Binding proteins and ase1/prc1 define local functionality of structurally distinct parts of the microtubule cytoskeleton. Trends Cell. Biol 2013, 23, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Kosetsu, K.; de Keijzer, J.; Janson, M.E.; Goshima, G. Microtubule-associated protein65 is essential for maintenance of phragmoplast bipolarity and formation of the cell plate in physcomitrella patens. Plant Cell 2013, 25, 4479–4492. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.; Hotta, T.; Guo, F.; Roberson, R.W.; Lee, Y.R.; Liu, B. Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein map65–3 in arabidopsis. Plant Cell 2011, 23, 2909–2923. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Smertenko, A.; Wagner, V.; Heinrich, M.; Hussey, P.J.; Hauser, M.T. The plant microtubule-associated protein atmap65–3/ple is essential for cytokinetic phragmoplast function. Curr. Biol. 2004, 14, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Sasabe, M.; Kosetsu, K.; Hidaka, M.; Murase, A.; Machida, Y. Arabidopsis thaliana map65–1 and map65–2 function redundantly with map65–3/pleiade in cytokinesis downstream of mpk4. Plant Signal Behav. 2011, 6, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Hiwatashi, Y.; Obara, M.; Sato, Y.; Fujita, T.; Murata, T.; Hasebe, M. Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss physcomitrella patens. Plant Cell 2008, 20, 3094–3106. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Carmena, M. Polo-Like kinase-activating kinases: Aurora a, aurora b and what else? Cell Cycle 2012, 11, 1490–1495. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.K.; Ozlu, N.; Coughlin, M.; Steen, J.J.; Mitchison, T.J. Plk1 negatively regulates prc1 to prevent premature midzone formation before cytokinesis. Mol. Biol. Cell 2012, 23, 2702–2711. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, D.; De Rybel, B.; Gudesblat, G.; Demidov, D.; Grunewald, W.; De Smet, I.; Houben, A.; Beeckman, T.; Russinova, E. Arabidopsis alpha aurora kinases function in formative cell division plane orientation. Plant Cell 2011, 23, 4013–4024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasabe, M.; Machida, Y. Regulation of organization and function of microtubules by the mitogen-activated protein kinase cascade during plant cytokinesis. Cytoskeleton 2012, 69, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Sasabe, M.; Soyano, T.; Takahashi, Y.; Sonobe, S.; Igarashi, H.; Itoh, T.J.; Hidaka, M.; Machida, Y. Phosphorylation of ntmap65–1 by a map kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev. 2006, 20, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Panteris, E.; Adamakis, I.D.; Voulgari, G.; Papadopoulou, G. A role for katanin in plant cell division: Microtubule organization in dividing root cells of fra2 and lue1arabidopsis thaliana mutants. Cytoskeleton 2011, 68, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Sano, T.; Sasabe, M.; Nonaka, S.; Higashiyama, T.; Hasezawa, S.; Machida, Y.; Hasebe, M. Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Collonnier, C.; Epert, A.; Mara, K.; Maclot, F.; Guyon-Debast, A.; Charlot, F.; White, C.; Schaefer, D.G.; Nogue, F. Crispr-cas9-mediated efficient directed mutagenesis and rad51-dependent and rad51-independent gene targeting in the moss physcomitrella patens. Plant Biotechnol. J. 2016. [Google Scholar] [CrossRef]
- Sugano, S.S.; Shirakawa, M.; Takagi, J.; Matsuda, Y.; Shimada, T.; Hara-Nishimura, I.; Kohchi, T. Crispr/cas9-mediated targeted mutagenesis in the liverwort marchantia polymorpha l. Plant Cell. Physiol. 2014, 55, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, K.; Nishihama, R.; Yamato, K.T.; Kohchi, T. Molecular genetic tools and techniques for marchantia polymorpha research. Plant Cell Physiol. 2016, 57, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, Y.; Kimura, A.; Tani, T.; Goshima, G. Cytoplasmic nucleation and atypical branching nucleation generate endoplasmic microtubules in physcomitrella patens. Plant Cell 2015, 27, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Kiyomitsu, T. Mechanisms of daughter cell-size control during cell division. Trends Cell Biol. 2015, 25, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, H.; Zachgo, S. The evolution of cell division: From streptophyte algae to land plants. Trends Plant Sci. 2016, 21, 872–883. [Google Scholar] [CrossRef] [PubMed]
| Generic Name | H. sapiens | D. melanogaster | S. pombe | P. patens | A. thaliana (* identified with BLAST) | A. thaliana Gene Acession # | References | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kinase/phosphatase/signalling | Cdk1 | + | + | + | * | AT3G48750 | ||||||||||||
| Aurora kinase | + | + | + | + | + | * | AT4G32830 etc. | [48] | ||||||||||
| Plk1 | + | + | + | [49] | ||||||||||||||
| Haspin | + | + | + | + | + | * | AT1G09450 | |||||||||||
| Ran | + | + | + | + | + | * | AT5G20010 etc. | |||||||||||
| RCC1 | + | + | + | + | + | * | AT5G63860 etc. | |||||||||||
| RanGAP | + | + | + | + | + | * | AT3G63130, AT5G19320 | |||||||||||
| PP2A | + | + | + | + | + | * | AT1G69960 etc. | [50] | ||||||||||
| Endosulfine | + | + | + | + | + | AT1G69510 | [51] | |||||||||||
| PP1 | + | + | + | + | + | AT2G29400 | [50] | |||||||||||
| PP6 | + | + | + | + | + | * | AT3G19980 | |||||||||||
| Centriole | Plk4 | + | + | [52] | ||||||||||||||
| Sas4 | + | + | ||||||||||||||||
| Sas5/Ana2/STIL | + | + | ||||||||||||||||
| Sas6 | + | + | [52] | |||||||||||||||
| Spd2/CEP192 | + | + | ||||||||||||||||
| Ana1/CEP295 | + | + | ||||||||||||||||
| Ana3/Rotatin | + | + | ||||||||||||||||
| Motor/MAPs | Kin4/chromokinesin | + | + | + | + | * | AT5G60930 etc. | [53,54] | ||||||||||
| Kin5 | + | + | + | + | + | * | AT2G28620 etc. | |||||||||||
| Kin6 | + | + | ||||||||||||||||
| Kin7/CENP-E | + | + | + | + | * | AT3G10180 etc. | ||||||||||||
| Kin8 | + | + | + | + | + | * | AT1G18550, AT3G49650 | |||||||||||
| Kin12/KIF15 | + | + | + | * | AT3G19050 etc. | |||||||||||||
| Kin13 | + | + | + | + | * | AT3G16060, AT3G16630 | ||||||||||||
| Kin14 | + | + | + | + | + | * | AT4G21270 etc. | |||||||||||
| DHC | + | + | + | [55] | ||||||||||||||
| DIC | + | + | + | |||||||||||||||
| DLC (LC8) | + | + | + | + | + | * | AT4G15930 etc. | |||||||||||
| Dynactin p50 | + | + | + | |||||||||||||||
| Dynactin p150 | + | + | + | |||||||||||||||
| Dynactin ARP1 | + | + | + | [56] | ||||||||||||||
| PRC1/MAP65/Ase1 | + | + | + | + | + | * | AT3G60840 etc. | |||||||||||
| Katanin (p60) | * | AT1G80350 | ||||||||||||||||
| HURP | + | + | ||||||||||||||||
| TACC | + | + | + | |||||||||||||||
| TPX2 | + | + | + | + | * | AT1G03780 etc. | [57] | |||||||||||
| Nucleation | γ-Tubulin | + | + | + | + | + | * | AT3G61650, AT5G05620 | [58] | |||||||||
| GCP2/3 | + | + | + | + | + | * | AT5G17410, AT5G06680 | [46] | ||||||||||
| GCP4/5/6 | + | + | + | + | + | * | At3g53760 etc. | [46,59] | ||||||||||
| NEDD1 | + | + | + | + | * | AT5G05970 | [60] | |||||||||||
| Mzt1 | + | + | + | + | + | * | AT1G73790, AT4G09550 | [61,62,63] | ||||||||||
| Mzt2 | + | [64] | ||||||||||||||||
| Augmin (8 subunits) | + | + | + | + | * | At5g40740 etc. | [17,46,65] | |||||||||||
| Pericentrin/D-plp | + | + | [64] | |||||||||||||||
| AKAP9 | + | [64,66] | ||||||||||||||||
| SPC110/Pcp1 | + | [64] | ||||||||||||||||
| CDK5RAP2/Cnn | + | + | + | |||||||||||||||
| Myomegalin | + | |||||||||||||||||
| Microtubule plus end | ch-TOG/XMAP215 | + | + | + | + | + | * | AT2G35630 | [67] | |||||||||
| EB1 | + | + | + | + | + | * | AT5G62500 etc. | |||||||||||
| SLAIN/Sentin | + | + | ||||||||||||||||
| CLIP170 | + | + | + | |||||||||||||||
| CLASP | + | + | + | + | + | * | AT2G20190 | |||||||||||
| SKAP | + | |||||||||||||||||
| Astrin | + | |||||||||||||||||
| Microtubule minus end | CAMSAP | + | + | [68] | ||||||||||||||
| Msd1/SSX2IP | + | + | * | AT5G57410 etc. | [69] | |||||||||||||
| ASPM | + | + | + | + | * | AT4G21820 | ||||||||||||
| CaM | + | + | + | + | + | * | AT2G27030 etc. | |||||||||||
| NuMA | + | + | ||||||||||||||||
| Microspherule | + | + | + | + | * | AT3G54350 etc. | ||||||||||||
| Chromosome | CAP-D2 | + | + | + | + | + | * | AT3G57060 | [70] | |||||||||
| SMC2 | + | + | + | + | + | * | AT3G47460, AT5G62410 | [70,71] | ||||||||||
| CAP-H | + | + | + | + | + | * | AT2G32590 | [70,72] | ||||||||||
| SMC4 | + | + | + | + | + | * | AT5G48600 | [70,71,72,73] | ||||||||||
| CAP-G | + | + | + | + | + | * | AT5G37630 | [70] | ||||||||||
| Topo II | + | + | + | + | + | * | AT3G23890 | |||||||||||
| Rad21 | + | + | + | + | + | * | AT5G16270 etc. | [74,75] | ||||||||||
| SCC3 | + | + | + | + | + | * | AT2G47980 | [75] | ||||||||||
| SMC1 | + | + | + | + | + | * | AT3G54670 | |||||||||||
| SMC3 | + | + | + | + | + | * | AT2G27170 | |||||||||||
| SCC2 | + | + | + | + | + | * | AT5G15540 | [76] | ||||||||||
| SCC4 | + | + | + | + | + | * | AT5G51340 | |||||||||||
| Eco1 | + | + | + | + | + | * | AT4G31400 | [77] | ||||||||||
| Sororin | + | + | [78] | |||||||||||||||
| Wapl | + | + | + | + | + | * | AT1G11060 | |||||||||||
| PDS5 | + | + | + | + | + | * | AT5G47690 etc. | |||||||||||
| Kinetochore/centromere | HP1 | + | + | + | + | + | * | AT5G17690 | ||||||||||
| Sgo1 | + | + | + | + | + | AT3G10440, AT5G04320 | [79] | |||||||||||
| Borealin | + | + | + | * | AT4g39630 | |||||||||||||
| INCENP | + | + | + | + | + | AT5g55820 | [80] | |||||||||||
| Survivin | + | + | + | |||||||||||||||
| CENP-B | + | + | ||||||||||||||||
| Mis18 | + | + | ||||||||||||||||
| Mis18BP1 | + | + | + | + | At5g02520 | [81] | ||||||||||||
| HJURP | + | + | ||||||||||||||||
| Cal1 | + | |||||||||||||||||
| CENP-A | + | + | + | + | + | * | AT1G01370 | [82] | ||||||||||
| CENP-C | + | + | + | + | + | * | AT1G15660 | [83] | ||||||||||
| CENP-S | + | + | + | + | * | AT5G50930 | [84] | |||||||||||
| CENP-X | + | + | + | + | AT1G78790 | [85] | ||||||||||||
| CENP-T | + | + | ||||||||||||||||
| CENP-W | + | + | ||||||||||||||||
| CENP-L | + | + | ||||||||||||||||
| CENP-N | + | + | ||||||||||||||||
| CENP-H | + | + | ||||||||||||||||
| CENP-I | + | ; | ||||||||||||||||
| CENP-K | + | + | ||||||||||||||||
| CENP-M | + | |||||||||||||||||
| CENP-O | + | + | + | + | * | AT5G10710 | ||||||||||||
| CENP-P | + | + | ||||||||||||||||
| CENP-Q | + | + | ||||||||||||||||
| CENP-U | + | + | ||||||||||||||||
| CENP-R | + | |||||||||||||||||
| Mis12 | + | + | + | + | + | AT5G35520 | [86] | |||||||||||
| Dsn1/Mis13 | + | + | + | AT3G27520 | ||||||||||||||
| Nnf1 | + | + | + | + | AT4G19350 | |||||||||||||
| Nsl1/Mis14 | + | + | + | |||||||||||||||
| KNL1 | + | + | + | AT2G04235 | [87] | |||||||||||||
| Ndc80 | + | + | + | + | + | * | AT3G54630 | |||||||||||
| Nuf2 | + | + | + | + | + | * | AT1G61000 | |||||||||||
| Spc24 | + | ? | + | + | + | AT3G08880, AT5G01570 | ||||||||||||
| Spc25 | + | + | + | + | + | * | AT3G48210 | |||||||||||
| Ska1 | + | + | * | AT3G60660 | ||||||||||||||
| Ska2 | AT2G24970 | |||||||||||||||||
| Ska3 | + | + | + | AT5G06590 | ||||||||||||||
| Dam1 | + | |||||||||||||||||
| CENP-F | + | |||||||||||||||||
| Spindle assembly checkpoint (SAC) | Mad1 | + | + | + | + | + | * | AT5G49880 | ||||||||||
| Mad2 | + | + | + | + | + | * | AT3G25980 | [88] | ||||||||||
| Mad3 /BubR1 | + | + | + | + | + | * | AT2G33560, AT5G05510 | [88,89] | ||||||||||
| Bub1 | + | + | + | + | + | * | AT2G20635 | |||||||||||
| Bub3 | + | + | + | + | + | * | AT3G19590, AT1G49910 | |||||||||||
| Mps1 | + | + | + | + | + | * | AT1G77720 | [90] | ||||||||||
| Tpr | + | + | + | + | + | * | AT1G79280 | [91] | ||||||||||
| Cdc20 | + | + | + | + | + | * | AT4G33270 etc. | |||||||||||
| Spindly | + | + | ||||||||||||||||
| Rod | + | + | ||||||||||||||||
| Zwilch | + | + | ||||||||||||||||
| Zw10 | + | + | + | + | * | AT2G32900 | [92] | |||||||||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, M.; Goshima, G. Mitotic Spindle Assembly in Land Plants: Molecules and Mechanisms. Biology 2017, 6, 6. https://doi.org/10.3390/biology6010006
Yamada M, Goshima G. Mitotic Spindle Assembly in Land Plants: Molecules and Mechanisms. Biology. 2017; 6(1):6. https://doi.org/10.3390/biology6010006
Chicago/Turabian StyleYamada, Moé, and Gohta Goshima. 2017. "Mitotic Spindle Assembly in Land Plants: Molecules and Mechanisms" Biology 6, no. 1: 6. https://doi.org/10.3390/biology6010006
APA StyleYamada, M., & Goshima, G. (2017). Mitotic Spindle Assembly in Land Plants: Molecules and Mechanisms. Biology, 6(1), 6. https://doi.org/10.3390/biology6010006





