Exploring the Glycans of Euglena gracilis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth of Euglena
2.2. Lectin Labelling
2.3. Sugar Nucleotide Profiling
2.4. Immunocarbohydrate Microarray Profiling
2.5. N- and O-Glycan Analysis
2.5.1. Sample Preparation
2.5.2. Mass Spectrometric Analysis
3. Results and Discussion
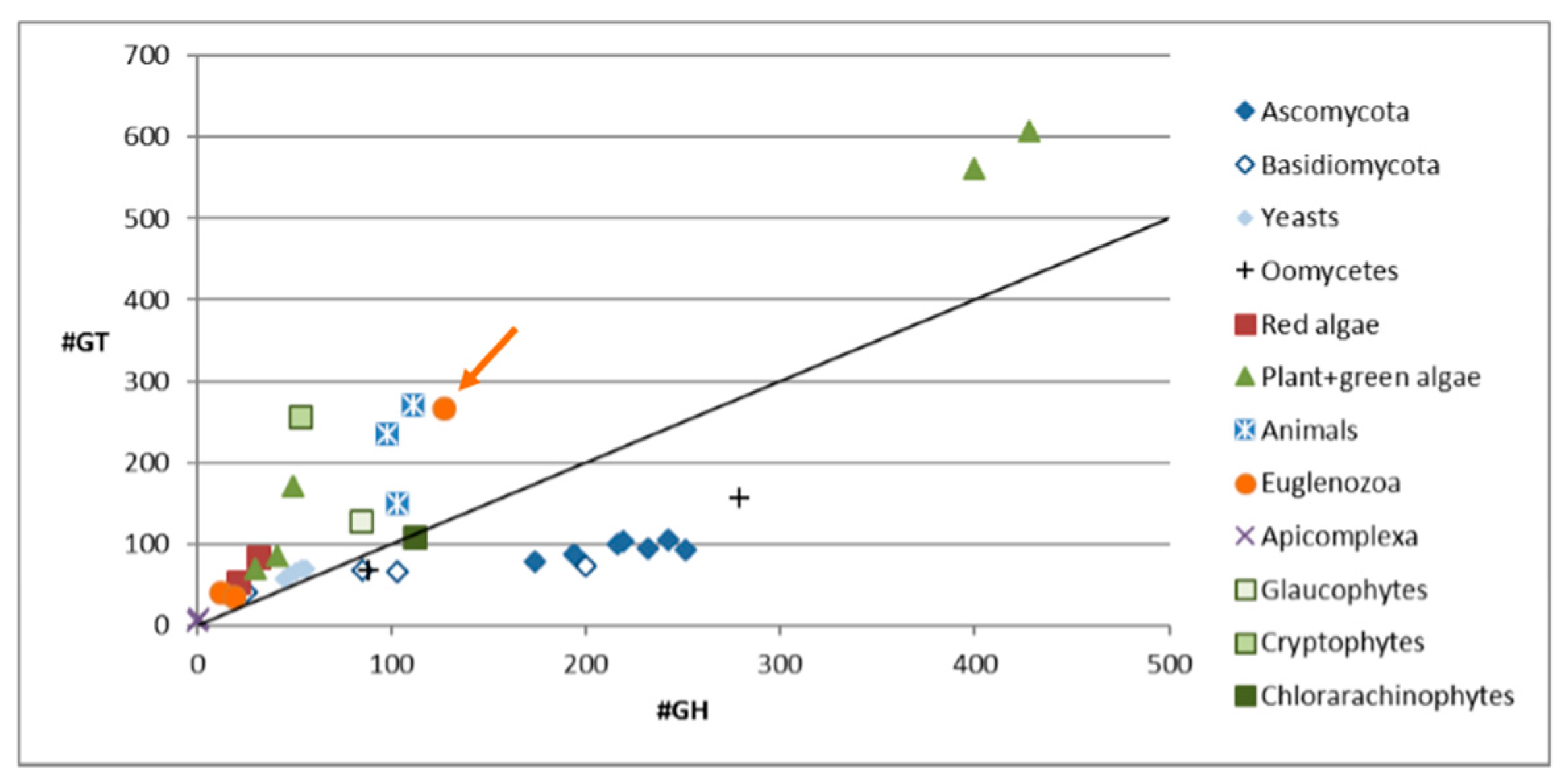
3.1. Carbohydrate Active Enzymes in Euglena
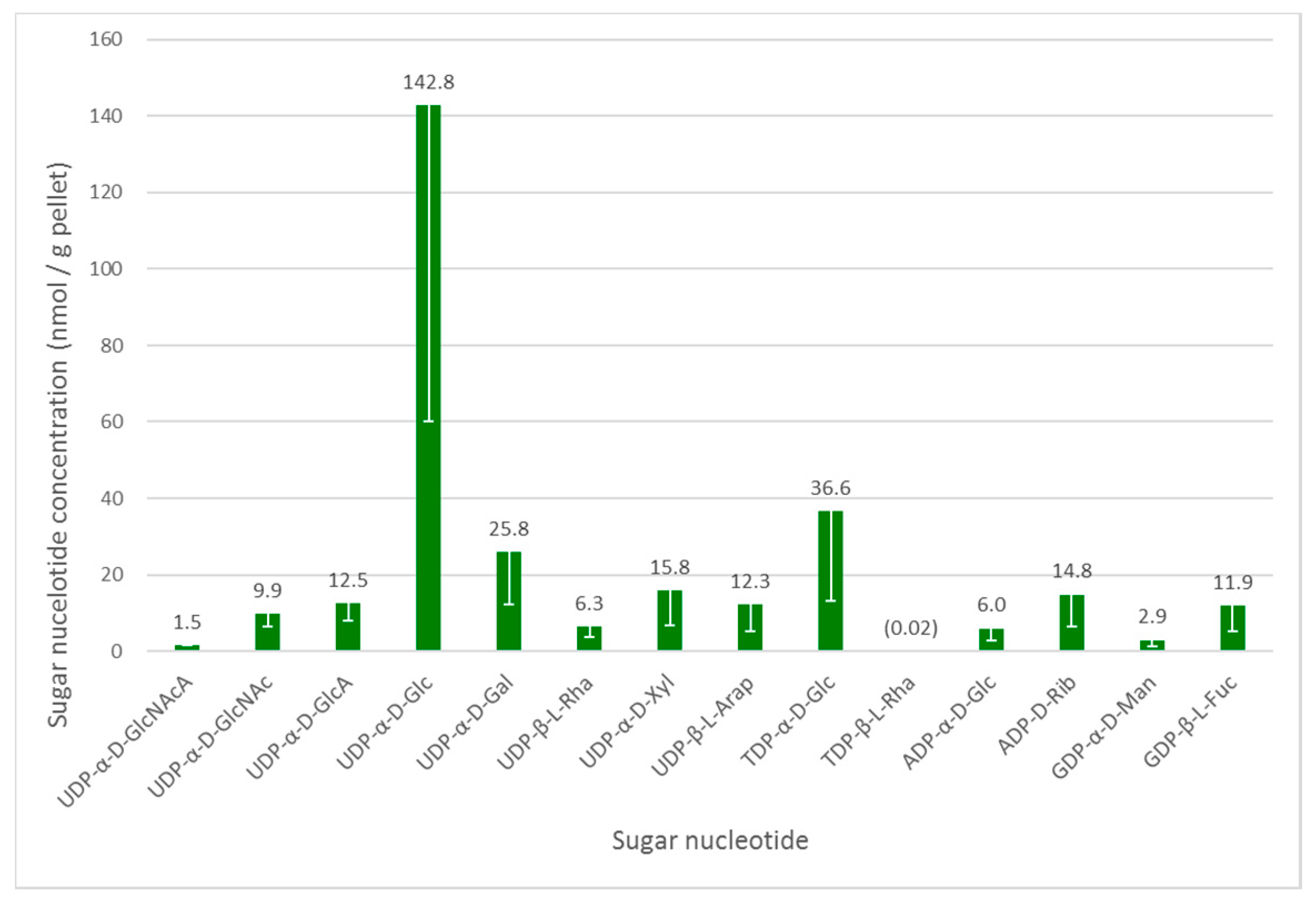
3.2. Sugar Nucleotide Profiling in Euglena
3.3. Glycan Analysis of Euglena
3.3.1. Surface Glycans
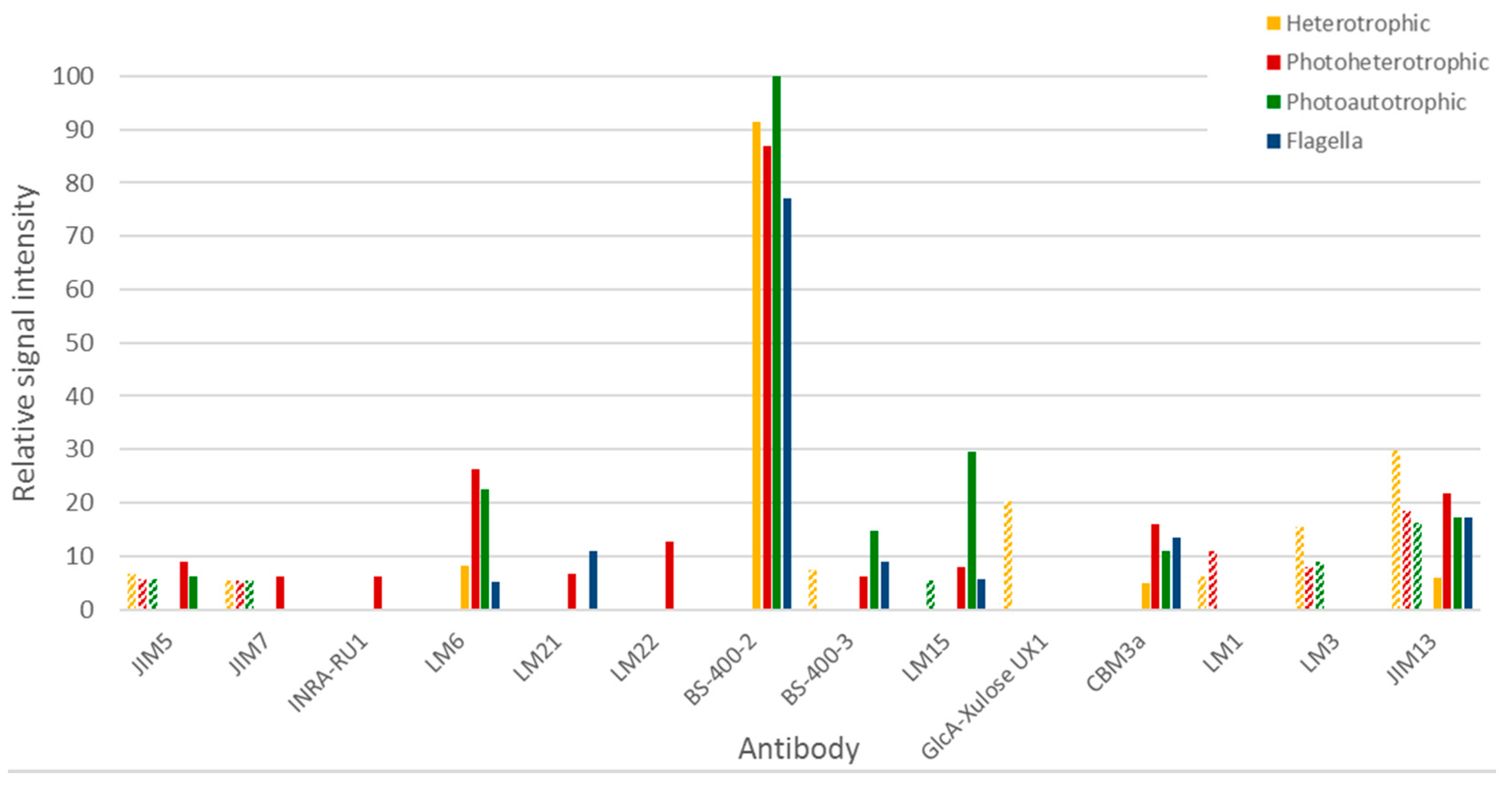
3.3.2. Immunocarbohydrate Microarray Profiling
3.3.3. Analysis of Euglena Protein Bound Glycans
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schwartzbach, S.; Shigeoka, S. Euglena: Biochemistry, Cell and Molecular Biology; Springer: Berlin, Germany, 2017. [Google Scholar]
- Krajčovič, J.; Matej, V.; Schwartzbach, S.D. Euglenoid flagellates: A multifaceted biotechnology platform. J. Biotechnol. 2015, 202, 135–145. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E.C.; Saalbach, G.; Field, R.A. Gene discovery for synthetic biology: Exploring the novel natural product biosynthetic capacity of eukaryotic microalgae. Methods Enzymol. 2016, 576, 99–120. [Google Scholar] [PubMed]
- O’Neill, E.C.; Trick, M.; Hill, L.; Rejzek, M.; Dusi, R.G.; Hamilton, C.J.; Zimba, P.V.; Henrissat, B.; Field, R.A. The transcriptome of Euglena gracilis reveals unexpected metabolic capabilities for carbohydrate and natural product biochemistry. Mol. Biosyst. 2015, 11, 2808–2820. [Google Scholar] [CrossRef] [PubMed]
- Barras, D.R.; Stone, B.A. Chemical composition of pellicle of Euglena gracilis var. bacillaris. Biochem. J. 1965, 97, 14–15. [Google Scholar]
- Jahn, T.L. Manual of Phycology: An Introduction to the Algae and Their Biology; Chronica Botanica Co.: Leyden, The Netherlands, 1951. [Google Scholar]
- Ivanova, I.M.; Nepogodiev, S.A.; Saalbach, G.; O’Neill, E.C.; Urbaniak, M.D.; Ferguson, M.A.J.; Gurcha, S.S.; Besra, G.S.; Field, R.A. Fluorescent mannosides serve as acceptor substrates for glycosyltransferase and sugar-1-phosphate transferase activities in Euglena gracilis membranes. Carbohydr. Res. 2017, 438, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Bouck, G.B.; Rogalski, A.; Valaitis, A. Surface organization and composition of Euglena. 2. Flagellar mastigonemes. J. Cell Biol. 1978, 77, 805–826. [Google Scholar] [CrossRef] [PubMed]
- De la Canal, L.; Parodi, A.J. Glycosylation of proteins in the protozoan Euglena gracilis. Comp. Biochem. Physiol. Part B Comp. Biochem. 1985, 81, 803–805. [Google Scholar] [CrossRef]
- Izquierdo, L.; Schulz, B.L.; Rodrigues, J.A.; Güther, M.L.S.; Procter, J.B.; Barton, G.J.; Aebi, M.; Ferguson, M.A.J. Distinct donor and acceptor specificities of Trypanosoma brucei oligosaccharyltransferases. EMBO J. 2009, 28, 2650–2661. [Google Scholar] [CrossRef] [PubMed]
- Geetha-Habib, M.; Bouck, G.B. Synthesis and mobilization of flagellar glycoproteins during regeneration in Euglena. J. Cell Biol. 1982, 93, 432–441. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E.C.; Trick, M.; Henrissat, B.; Field, R.A. Euglena in time: Evolution, control of central metabolic processes and multi-domain proteins in carbohydrate and natural product biochemistry. Perspect. Sci. 2015, 6, 84–93. [Google Scholar] [CrossRef]
- Olszewski, N.E.; West, C.M.; Sassi, S.O.; Hartweck, L.M. O-GlcNAc protein modification in plants: Evolution and function. Biochim. Biophys. Acta 2010, 1800, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Zentella, R.; Sui, N.; Barnhill, B.; Hsieh, W.-P.; Hu, J.; Shabanowitz, J.; Boyce, M.; Olszewski, N.E.; Zhou, P.; Hunt, D.F.; et al. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol. 2017, 13, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Rejzek, M.; Hill, L.; Hems, E.S.; Kuhaudomlarp, S.; Wagstaff, B.A.; Field, R.A. Chapter Seven—Profiling of sugar nucleotides. Methods Enzymol. 2017, 597, 209–238. [Google Scholar] [PubMed]
- Wagstaff, B.A.; Rejzek, M.; Kuhaudomlarp, S.; Hill, L.; Mascia, I.; Nepogodiev, S.A.; Field, R.A. NDP-β-L-rhamnose biosynthesis across the algal taxonomic groups; an evolutionary perspective. (under review).
- Turnock, D.C.; Ferguson, M.A.J. Sugar nucleotide pools of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Eukaryot. Cell 2007, 6, 1450–1463. [Google Scholar] [CrossRef] [PubMed]
- Räbinä, J.; Mäki, M.; Savilahti, E.M.; Järvinen, N.; Penttilä, L.; Renkonen, R. Analysis of nucleotide sugars from cell lysates by ion-pair solid-phase extraction and reversed-phase high-performance liquid chromatography. Glycoconj. J. 2001, 18, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Pabst, M.; Grass, J.; Fischl, R.; Léonard, R.; Jin, C.; Hinterkörner, G.; Borth, N.; Altmann, F. Nucleotide and nucleotide sugar analysis by liquid chromatography-electrospray ionization-mass spectrometry on surface-conditioned porous graphitic carbon. Anal. Chem. 2010, 82, 9782–9788. [Google Scholar] [CrossRef] [PubMed]
- Rejzek, M.; Mukhopadhyay, B.; Wenzel, C.Q.; Lam, J.S.; Field, R.A. Direct oxidation of sugar nucleotides to the corresponding uronic acids: TEMPO and platinum-based procedures. Carbohydr. Res. 2007, 342, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Errey, J.C.; Mann, M.C.; Fairhurst, S.A.; Hill, L.; McNeil, M.R.; Naismith, J.H.; Percy, J.M.; Whitfield, C.; Field, R.A. Sugar nucleotide recognition by Klebsiella pneumoniae UDP-d-galactopyranose mutase: Fluorinated substrates, kinetics and equilibria. Org. Biomol. Chem. 2009, 7, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Errey, J.C.; Mukhopadhyay, B.; Kartha, K.P.R.; Field, R.A. Flexible enzymatic and chemo-enzymatic approaches to a broad range of uridine-diphospho-sugars. Chem. Commun. 2004, 2706–2707. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.; Sorensen, I.; Bernal, A.J.; Blaukopf, C.; Lee, K.; Obro, J.; Pettolino, F.; Roberts, A.; Mikkelsen, J.D.; Knox, J.P.; et al. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 2007, 50, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.L.; Fangel, J.U.; McCleary, B.; Ruzanski, C.; Rydahl, M.G.; Ralet, M.-C.; Farkas, V.; von Schantz, L.; Marcos, S.E.; Andersen, M.C.F.; et al. Versatile high-resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 2012, 287, 39429–39438. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.H.; Karlsson, N.G.; Kolarich, D.; Packer, N.H. Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 2012, 7, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Zavala, J.S.; Ortiz-Cruz, M.A.; Mendoza-Hernández, G.; Moreno-Sánchez, R. Increased synthesis of α-tocopherol, paramylon and tyrosine by Euglena gracilis under conditions of high biomass production. J. Appl. Microbiol. 2010, 109, 2160–2172. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.A. Chemistry, Biochemistry, and Biology of 1–3 Beta Glucans and Related Polysaccharides; Academic Press: San Diego, CA, USA, 2009. [Google Scholar]
- Freeze, H.H.; Hart, G.W.; Schnaar, R.L. Glycosylation precursors. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017. [Google Scholar]
- Qu, Q.; Lee, S.J.; Boos, W. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 2004, 279, 47890–47897. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, X. ADP-Ribosyltransferases and Poly ADP-Ribosylation. Curr. Protein Pept. Sci. 2015, 16, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Queval, G.; Gakière, B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 2006, 57, 1603–1620. [Google Scholar] [CrossRef] [PubMed]
- So, L.L.; Goldstein, I.J. Protein-carbohydrate interaction XX. On the number of combining sites of concanavalin A, the phytohemagglutinin of the jack bean. Biochim. Biophys. Acta (BBA) Gen. Subj. 1968, 165, 398–404. [Google Scholar] [CrossRef]
- Hard, K.; Van Doorn, J.M.; Thomas-Oates, J.E.; Kamerling, J.P.; Van der Horst, D.J. Structure of the Asn-linked oligosaccharides of apolipophorin III from the insect Locusta migratoria. Carbohydrate-linked 2-aminoethylphosphonate as a constituent of a glycoprotein. Biochemistry 1993, 32, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Eckmair, B.; Jin, C.; Abed-Navandi, D.; Paschinger, K. Multistep fractionation and mass dpectrometry reveal zwitterionic and anionic modifications of the N- and O-glycans of a marine snail. Mol. Cell. Proteom. 2016, 15, 573–597. [Google Scholar] [CrossRef] [PubMed]
- Mathieu-Rivet, E.; Scholz, M.; Arias, C.; Dardelle, F.; Schulze, S.; Le Mauff, F.; Teo, G.; Hochmal, A.K.; Blanco-Rivero, A.; Loutelier-Bourhis, C.; et al. Exploring the N-glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol. Cell. Proteom. 2013, 12, 3160–3183. [Google Scholar] [CrossRef] [PubMed]
- Zamze, S.E.; Ashford, D.A.; Wooten, E.W.; Rademacher, T.W.; Dwek, R.A. Structural characterization of the asparagine-linked oligosaccharides from Trypanosoma brucei type II and type III variant surface glycoproteins. J. Biol. Chem. 1991, 266, 20244–20261. [Google Scholar] [PubMed]
- Jones, D.C.; Mehlert, A.; Guther, M.L.; Ferguson, M.A. Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J. Biol. Chem. 2005, 280, 35929–35942. [Google Scholar] [CrossRef] [PubMed]
- Deeg, M.A.; Humphrey, D.R.; Yang, S.H.; Ferguson, T.R.; Reinhold, V.N.; Rosenberry, T.L. Glycan components in the glycoinositol phospholipid anchor of human erythrocyte acetylcholinesterase. Novel fragments produced by trifluoroacetic acid. J. Biol. Chem. 1992, 267, 18573–18580. [Google Scholar] [PubMed]
- Kallolimath, S.; Castilho, A.; Strasser, R.; Grunwald-Gruber, C.; Altmann, F.; Strubl, S.; Galuska, C.E.; Zlatina, K.; Galuska, S.P.; Werner, S.; et al. Engineering of complex protein sialylation in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 9498–9503. [Google Scholar] [CrossRef] [PubMed]

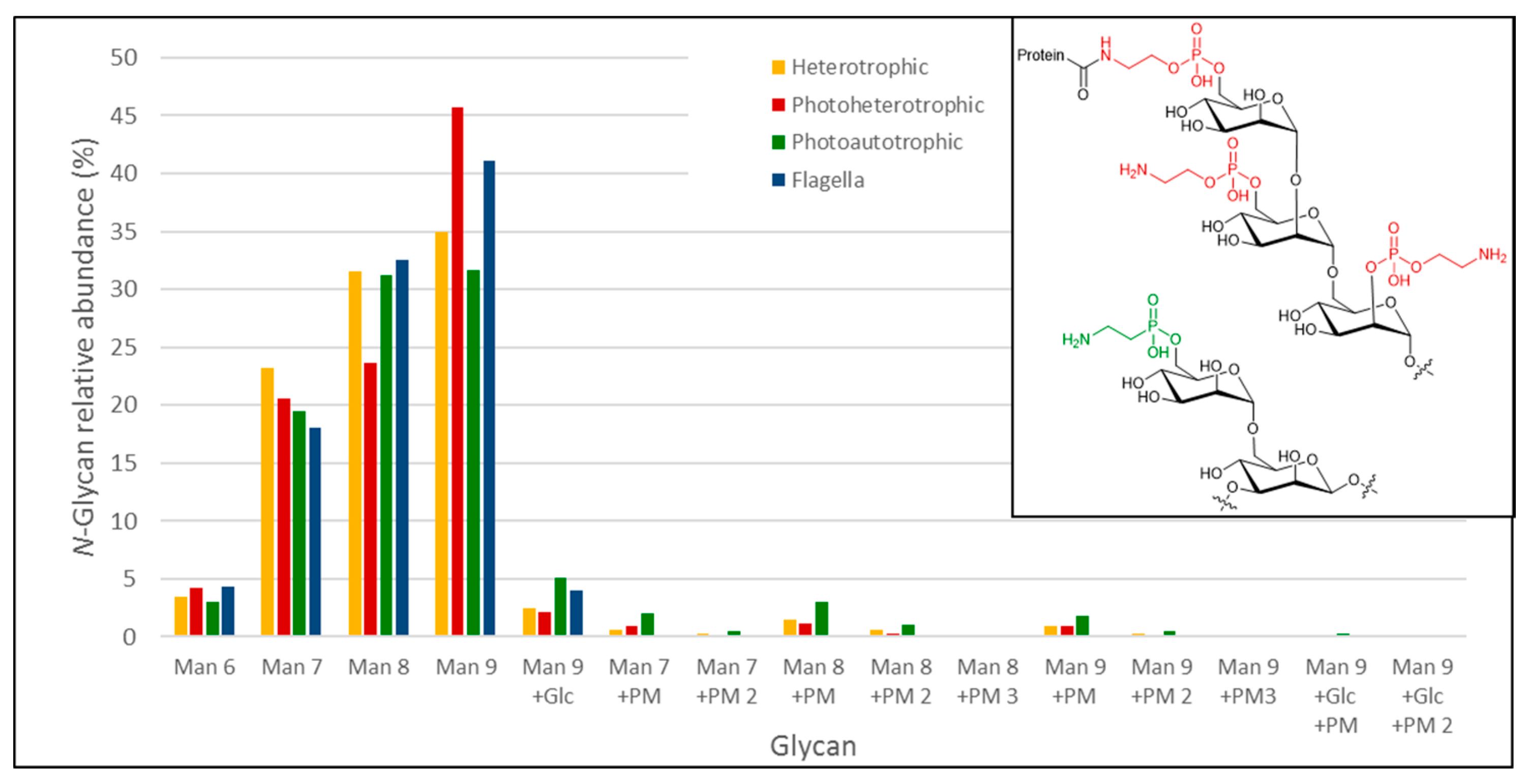
| Observed m/z | Charge State | Calculated Mass (Da) | TheoreticalMass (Da) | Delta Mass (Da) | Glycan Identified | |
|---|---|---|---|---|---|---|
| 698.24 | 2 | 1398.48 | 1398.5 | 0.02 | (Hex)3 + (Man)3(GlcNAc)2 | Man 6 |
| 779.25 | 2 | 1560.50 | 1560.55 | 0.05 | (Hex)4 + (Man)3(GlcNAc)2 | Man 7 |
| 860.26 | 2 | 1722.52 | 1722.60 | 0.08 | (Hex)5 + (Man)3(GlcNAc)2 | Man 8 |
| 941.37 | 2 | 1884.74 | 1884.66 | −0.08 | (Hex)6 + (Man)3(GlcNAc)2 | Man 9 |
| 1022.32 | 2 | 2046.64 | 2046.71 | 0.07 | (Hex)7 + (Man)3(GlcNAc)2 | Man 9 + Glc |
| 832.75 | 2 | 1667.50 | 1667.55 | 0.05 | (107)(Hex)4 + (Man)3(GlcNAc)2 | Man 7 + PM |
| 886.25 | 2 | 1774.50 | 1774.55 | 0.05 | (107)2(Hex)4 + (Man)3(GlcNAc)2 | Man 7 + PM 2 |
| 913.76 | 2 | 1829.52 | 1829.60 | 0.08 | (107)(Hex)5 + (Man)3(GlcNAc)2 | Man 8 + PM |
| 967.26 | 2 | 1936.52 | 1936.60 | 0.08 | (107)2(Hex)5 + (Man)3(GlcNAc)2 | Man 8 + PM 2 |
| 1020.76 | 2 | 2043.52 | 2043.60 | 0.08 | (107)3(Hex)5 + (Man)3(GlcNAc)2 | Man 8 + PM 3 |
| 994.87 | 2 | 1991.74 | 1991.66 | −0.08 | (107)(Hex)6 + (Man)3(GlcNAc)2 | Man 9 + PM |
| 1048.37 | 2 | 2098.74 | 2098.66 | −0.08 | (107)2(Hex)6 + (Man)3(GlcNAc)2 | Man 9 + PM 2 |
| 1101.87 | 2 | 2205.74 | 2205.66 | −0.08 | (107)3(Hex)6 + (Man)3(GlcNAc)2 | Man 9 + PM 3 |
| 1075.82 | 2 | 2153.64 | 2153.71 | 0.07 | (107)(Hex)7 + (Man)3(GlcNAc)2 | Man 9 + Glc + PM |
| 1129.32 | 2 | 2260.64 | 2260.71 | 0.07 | (107)2(Hex)7 + (Man)3(GlcNAc)2 | Man 9 + Glc + PM 2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Neill, E.C.; Kuhaudomlarp, S.; Rejzek, M.; Fangel, J.U.; Alagesan, K.; Kolarich, D.; Willats, W.G.T.; Field, R.A. Exploring the Glycans of Euglena gracilis. Biology 2017, 6, 45. https://doi.org/10.3390/biology6040045
O’Neill EC, Kuhaudomlarp S, Rejzek M, Fangel JU, Alagesan K, Kolarich D, Willats WGT, Field RA. Exploring the Glycans of Euglena gracilis. Biology. 2017; 6(4):45. https://doi.org/10.3390/biology6040045
Chicago/Turabian StyleO’Neill, Ellis C., Sakonwan Kuhaudomlarp, Martin Rejzek, Jonatan U. Fangel, Kathirvel Alagesan, Daniel Kolarich, William G. T. Willats, and Robert A. Field. 2017. "Exploring the Glycans of Euglena gracilis" Biology 6, no. 4: 45. https://doi.org/10.3390/biology6040045
APA StyleO’Neill, E. C., Kuhaudomlarp, S., Rejzek, M., Fangel, J. U., Alagesan, K., Kolarich, D., Willats, W. G. T., & Field, R. A. (2017). Exploring the Glycans of Euglena gracilis. Biology, 6(4), 45. https://doi.org/10.3390/biology6040045








