Was the Watchmaker Blind? Or Was She One-Eyed?
Abstract
:1. Introduction
1.1. Background
“Life isn’t like that. Evolution has no long-term goal. There is no long-distance target, no final perfection to serve as a criterion for selection, although human vanity cherishes the absurd notion that our species is the final goal of evolution. In real life, the criterion for selection is always short-term, either simple survival or, more generally, reproductive success.”
1.2. Purpose and Organization of This Article
2. Definitions
3. Goals within Organisms
3.1. Regulated (Directed) Hypermutation Processes
3.2. Is the System Purposive?
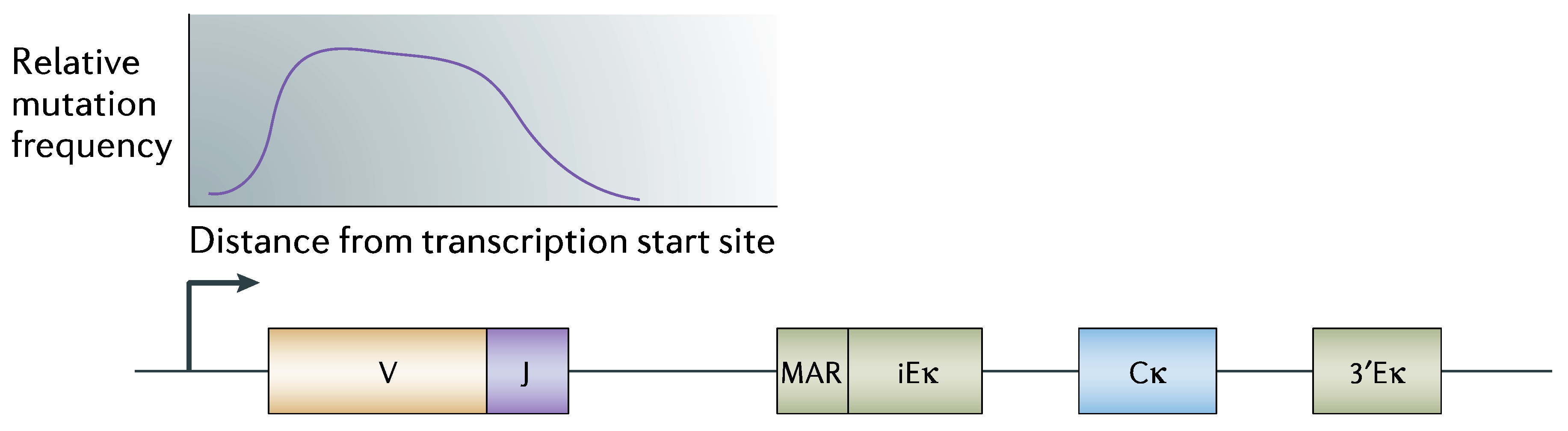
“Somatic hypermutation (SHM) introduces mutations in the variable region of immunoglobulin genes at a rate of ~10−3 mutations per base pair per cell division, which is 106-fold higher than the spontaneous mutation rate in somatic cells. To ensure genomic integrity, SHM needs to be targeted specifically to immunoglobulin genes.”
3.3. Natural Genetic Engineering
“In the future attention undoubtedly will be centered on the genome, and with greater appreciation of its significance as a highly sensitive organ of the cell, monitoring genomic activities and correcting common errors, sensing the unusual and unexpected events, and responding to them, often by restructuring the genome. We know about the components of genomes that could be made available for such restructuring. We know nothing, however, about how the cell senses danger and instigates responses to it that often are truly remarkable” (our italics).[34]
4. Goals within Populations
4.1. Contingency Loci in Bacteria
“This phenotypic variation, which is stochastic with respect to the timing of switching but has a programmed genomic location, allows a large repertoire of phenotypic solutions to be explored, while minimizing deleterious effects on fitness.”.[37]
“The strategy of generating multiple mutant chromosomes within a single cell may represent a widespread and conserved mechanism for the rapid evolution of genome change in response to unfavorable environments (i.e., chemo-therapy drugs and antibiotics)”.[39]
“Signaling pathways that sense environmental nutrients control genome change at the ribosomal DNA. This demonstrates that not all genome changes occur at random and that cells possess specific mechanisms to optimize their genome in response to the environment.” (our italics).[40]
4.2. Genetic Buffering by Regulatory Networks
4.3. Switch of Function in Regulatory Networks
“Step one mutations increase intracellular levels of phosphorylated NtrC, a distant homolog of FleQ, which begins to commandeer control of the FleQ regulon at the cost of disrupting nitrogen uptake and assimilation. Step two is a switch-of-function mutation that redirects NtrC away from nitrogen uptake and toward its novel function as a flagellar regulator. Our results demonstrate that natural selection can rapidly rewire regulatory networks in very few, repeatable mutational steps”.
4.4. Roles of Stochasticity and Natural Selection
4.5. Communication to the Genome
5. Speculation 1: Goals Achieved through Epigenetic Inheritance
5.1. Different Forms of Epigenetics
5.2. Experimental Examples
5.3. Role in Speciation
“From the second generation onwards the lineage bred endogamously, and despite intense inbreeding, was ecologically successful and showed transgressive segregation of bill morphology. This example shows that reproductive isolation, which typically develops over hundreds of generations, can be established in only three.”.[60]
6. Speculation 2: The Evolution of Goal-Directed Behaviour
6.1. The Continuity of Animal and Human Evolution
“the picture of the external hand of natural selection doing all the work is so compelling that it is easy to regard organisms as if they were entirely passive in the evolutionary process.”.(p. 105)
6.2. The Adaptability Driver
“Charles Darwin (1871) argued that choice of a mate could drive evolution. He called the evolutionary process ‘sexual selection’. Alfred Russel Wallace, although the co-author with Darwin of the first clear statement about the role of Natural Selection, did not like the new idea. Indeed, for many years most biologists did not take sexual selection seriously. When I was an undergraduate I was told confidently that, even if it were possible in theory, the process probably played little part in biological evolution. In recent years, however, many experiments have supported Darwin’s thinking.”.[67]
6.3. The Role of Contextual Logic in the Behavior of Organisms
7. Conclusions
7.1. The Harnessing of Stochasticity
7.2. Organisms as Agents
‘Lamarck’s claim that … there is a radical difference between living beings and inanimate objects might lead people to think that he was a vitalist. But he is not. On the contrary, his biology is a mechanistic reply to the physiological vitalism of Bichat, which was then the dominant theory’.[86]. (Our translation of Pichot’s French)
Being scared of Lamarckism leads to the neglect of the evolutionary effects of evolved systems that allow the inheritance of targeted and acquired variations. When the fact that variation is highly constrained and is shaped (or, rather, drafted) by the rules of the generating system is ignored, evolution cannot be properly understood.[88]
7.3. Organisms and Their Populations Are the One-Eyed Watchmakers
Acknowledgements
Conflicts of Interest
References and Notes
- Dawkins, R. The Blind Watchmaker; Norton & Company: New York, NY, USA, 1986. [Google Scholar]
- The Time Required Would in Fact Require Billions More Periods of Time Equivalent to the Whole Duration of the Universe. Available online: https://en.wikipedia.org/wiki/Weasel_program (accessed on 6 July 2017).
- Natarajan, C.; Hoffmann, F.G.; Weber, R.E.; Fago, A.; Witt, C.C.; Storz, J.F. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science 2016, 354, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Hillenmeyer, M.E.; Fung, E.; Wildenhain, J.; Pierce, S.E.; Hoon, S.; Lee, W.; Proctor, M.; St Onge, R.P.; Tyers, M.; Koller, D.; et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science 2008, 320, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. A theory of biological relativity: No privileged level of causation. Interface Focus 2012, 2, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Dance to the Tune of Life: Biological Relativity; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Charlesworth, D.; Barton, N.H.; Charlesworth, B. The sources of adaptive variation. Proc. R. Soc. B 2017, 284, 20162864. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, R. The Selfish Gene; OUP: Oxford, UK, 1976, 2006. [Google Scholar]
- Coyne, J.A. Why Evolution is True; OUP: New York, NY, USA, 2010. [Google Scholar]
- Bateson, P. The active role of behaviour in evolution. Biol. Philos. 2004, 19, 283–298. [Google Scholar] [CrossRef]
- Noble, D. Evolution viewed from physics, physiology and medicine. Interface Focus 2017, 7, 20160159. [Google Scholar] [CrossRef] [PubMed]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Suel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Oettmeier, C.; Brix, K.; Döbereiner, H.-G. Physarum polycephalum—A new take on a classic model system. J. Phys. D 2017, 50, 413001. [Google Scholar] [CrossRef]
- Monod, J.; Jacob, F. Teleonomic mechanisms in cellular metabolism, growth and differentiation. Cold Spring Harb. Symp. Quant. Biol. 1961, 26, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Weismann, A. The Germ-Plasm: A Theory of Heredity; Charles Scribner’s Sons: New York, NY, USA, 1893. [Google Scholar]
- Weismann, A. Die Allmacht der Naturzüchtung; Eine Erwiderung an Herbert Spencer; Fischer: Jena, Germany, 1893. [Google Scholar]
- Spencer, H. The Inadequacy of “Natural Selection”; University of Michigan Library: Ann Arbor, MI, USA, 1893. [Google Scholar] Reprinted in The Principles of Biology; London, D. Appleton and Company: New York, NY, USA, 1897.
- Mitchell, P.C. The Spencer-Weismann Controversy. Nature 1894, 49, 373–374. [Google Scholar] [CrossRef]
- Winther, R.G. August Weismann on Germ-Plasm Variation. J. Hist. Biol. 2001, 34, 517–555. [Google Scholar] [CrossRef] [PubMed]
- Segerstrale, U. Neo-darwinism. In Encyclopedia of Evolution; Pagel, M., Ed.; Oxford University Press: Oxford, UK, 2002; Volume 2, pp. 807–810. [Google Scholar]
- Bodmer, W.; McKie, R. Orion: The Book of Man: The Quest to Discover Our Genetic Heritage; Scribner: New York, NY, USA, 1995. [Google Scholar]
- Odegard, V.H.; Schatz, D.G. Targeting of somatic hypermutation. Nat. Rev. Immunol. 2006, 8, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Jones, S. The Language of the Genes; Harper-Collins: London, UK, 2000. [Google Scholar]
- Shapiro, J.A. Evolution: A View from the 21st Century; Pearson Education Inc.: Upper Saddle River, NJ, USA, 2011. [Google Scholar]
- For a Relatively Simple Account of How All This Is Achieved. Available online: https://www.scientificamerican.com/article/how-do-white-blood-cells/ (accessed on 6 July 2017).
- Kimura, M. On the evolutionary adjustment of spontaneous mutation rates. Genet. Res. 1967, 9, 23–34. [Google Scholar] [CrossRef]
- Moxon, E.R.; Rainey, P.B.; Nowak, M.A.; Lenski, R.E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 1994, 4, 24–33. [Google Scholar] [CrossRef]
- Lynch, M. Evolution of the mutation rate. Trends Genet. 2010, 26, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, J. No gene-specific optimization of mutation rate in Escherichia coli. Mol. Biol. Evol. 2013, 30, 1559–1562. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Luscombe, N.M. Non-random mutation: The evolution of targeted hypermutation and hypomutation. BioEssays 2013, 35, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, D.M.; Hastings, P.J.; Rosenberg, S.M. Stress-induced mutagenesis: Implications in cancer and drug resistance. Ann. Rev. Cancer Biol. 2017, 1, 119–140. [Google Scholar] [CrossRef]
- Maynard Smith, J.; Szathmáry, E. The Major Transitions in Evolution; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Moxon, R.; Bayliss, C.; Hood, D. Bacterial contingency loci: The role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 2006, 40, 307–333. [Google Scholar] [CrossRef] [PubMed]
- Moxon, E.R.; Thaler, D.S. The tinkerer’s evolving tool-box. Nature 1997, 387, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Ritz, D.; Lim, J.; Reynolds, C.; Poole, L.; Beckwith, J. Conversion of a perixiredoxin into a disulphide reductase by a triplet repeat expansion. Science 2001, 294, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.; Zhang, Q.; Vyawahare, S.; Rogers, E.; Rosenberg, S.M.; Austin, R. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc. Natl. Acad. Sci. USA 2015, 112, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.V.; Cruz, C.; Hull, R.M.; Ralser, M.; Houseley, J. Regulation of ribosomal DNA amplification by the tor pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 9674–9679. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Neo-darwinism, the modern synthesis, and selfish genes: Are they of use in physiology? J. Physiol. 2011, 589, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Differential and integral views of genetics in computational systems biology. Interface Focus 2011, 1, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. The Strategy of the Genes; Allen and Unwin: London, UK, 1957. [Google Scholar]
- Callaway, E. Genome studies attract criticism. Nature 2017, 546, 463. [Google Scholar] [CrossRef]
- Boyle, E.A.; Li, Y.; Pritchard, J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Breen, M.S.; Kemena, C.; Vlasov, P.K.; Notredame, C.; Kondrashov, F.A. Epistasis as the primary factor in molecular evolution. Nature 2012, 490, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.B.; Mulley, G.; Dills, A.H.; Alsohim, A.S.; McGuffin, L.J.; Studholme, D.J.; Silby, M.W.; Brockhurst, M.A.; Johnson, L.J.; Jackson, R.W. Evolutionary resurrection of flagellar motility via rewiring of the nitrogen regulation system. Science 2015, 347, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Kar, P.; Mirams, G.R.; Christian, H.C.; Parekh, A.B. Control of nfat isoform activation and nfat-dependent gene expression through two coincident and spatially segregated intracellular Ca2+ signals. Mol. Cell 2016, 64, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Groth, R.D.; Cohen, S.M.; Emery, J.F.; Li, B.; Hoedt, E.; Zhang, G.; Neubert, T.A.; Tsien, R.W. Γcamkii shuttles Ca2+/CAM to the nucleus to trigger creb phosphorylation and gene expression. Cell 2014, 159, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Weaver, I.C.G. Life at the interface between a dynamic environment and a fixed genome. In Mammalian Brain Development; Janigro, D., Ed.; Humana Press, Springer: New York, NY, USA, 2009; pp. 17–40. [Google Scholar]
- Rechavi, O.; Minevish, G.; Hobert, O. Transgenerational inheritance of an acquired small RNA-based antiviral response in c. Elegans. Cell 2011, 147, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Tollefsbol, T. Transgenerational Epigenetics: Evidence and Debate; Academic Press: Waltham, MA, USA, 2014. [Google Scholar]
- Skinner, M.K.; Gurerrero-Bosagna, C.; Haque, M.M.; Nilsson, E.E.; Koops, J.A.H.; Knutie, S.A.; Clayton, D.H. Epigenetics and the evolution of darwin’s finches. Genome Biol. Evol. 2014, 6, 1972–1989. [Google Scholar] [CrossRef] [PubMed]
- Soen, Y.; Knafo, M.; Elgart, M. A principle of organization which facilitates broad lamarckian-like adaptations by improvisation. Biol. Direct 2015, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.J.; Spencer, H.G.; Donohue, K.; Sultan, S.E. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 2013, 68, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Weaver, I.C.G.; Cervoni, N.; Champagne, F.A.; D’Alessio, A.C.; Sharma, S.; Sekl, J.R.; Dymov, S.; Szyf, M.; Meaney, M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004, 7, 847–854. [Google Scholar]
- Hanson, M.; Skinner, M. Developmental origins of epigenetic transgenerational inheritance. Environ. Epigenet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Menger, F.M. Molecular lamarckism: On the evolution of human intelligence. World Futures 2017, 73, 89–103. [Google Scholar] [CrossRef]
- Skoblov, M.Y.; Scobeyeva, V.A.; Baranova, A.V. The mechanisms of transgenerational inheritance and their potential contribution to human phenotypes. Russ. J. Genet. 2016, 52, 249–256. [Google Scholar] [CrossRef]
- Lamichhaney, S.; Han, F.; Webster, M.T.; Andersson, L.; Grant, B.R.; Grant, P.R. Rapid hybrid speciation in darwin’s finches. Science 2017. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. Epigenetic control of mobile DNA as an interface between experience and genome change. Front. Genet. 2014, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Bateson, P. Behaviour, Development and Evolution; Open Book Publishers: Cambridge, UK, 2017. [Google Scholar]
- Curiously, neo-Darwinists sometimes seem to be forced to concede a view that humans are unique. Consider this statement from The Selfish Gene: “Let us try to teach generosity and altruism, because we are born selfish. Let us understand what our own selfish genes are up to, because we may then at least have the chance to upset their designs, something that no other species has ever aspired to.” (chapter 1, our emphasis).
- Brosnan, S.F.; De Waal, F.B. Monkeys reject unequal pay. Nature 2003, 425, 297–299. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, S.F. A hypothesis of the co-evolution of cooperation and responses to inequity. Front. Neurosci. 2011. [Google Scholar] [CrossRef] [PubMed]
- Essler, J.L.; Marshall-Pescini, S.; Range, F. Domestication does not explain the presence of inequity aversion in dogs. Curr. Biol. 2017, 27, 1861–1865. [Google Scholar] [CrossRef] [PubMed]
- Bateson, P. New thinking about biological evolution. Biol. J. Linnean Soc. 2013. [Google Scholar] [CrossRef]
- Available online: https://www.princeton.edu/~dir/ (accessed on 6 July 2017).
- Leimgruber, K.L.; Rosati, A.G.; Santos, L.R. Capuchin monkeys punish those who have more. Evol. Hum. Behav. 2016, 37, 236–244. [Google Scholar] [CrossRef]
- Bronfman, Z.Z.; Ginsburg, S.; Jablonka, E. The transition to minimal consciousness through the evolution of associative learning. Front. Psychol. 2016, 7, 1954. [Google Scholar] [CrossRef] [PubMed]
- Corning, P.A. Evolution ‘on purpose’: How behaviour has shaped the evolutionary process. Biol. J. Linnean Soc. 2014, 112, 242–260. [Google Scholar] [CrossRef]
- Darwin, C. The Descent of Man, and Selection in Relation to Sex; John Murray: London, UK, 1871. [Google Scholar]
- We have put ‘know’ in parentheses to indicate that this does not necessarily imply conscious knowledge. In a general sense of ‘know’ there can be no doubt that the word is correct. We as humans ‘know’ how to control our blood pressure even though we also know that this knowledge is unconscious).
- Baldwin, J. A new factor in evolution. Am. Nat. 1896, 30, 441–451. [Google Scholar] [CrossRef]
- Avital, E.; Jablonka, E. Animal Traditions. Behavioural Inheritance in Evolution; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Bateson, P. The adaptability driver: Links between behaviour and evolution. Biol. Theory 2006, 1, 342–345. [Google Scholar] [CrossRef]
- Spalding, D.A. Instinct. With original observations on young animals. Macmillan’s Mag. 1873, 27, 282–293. [Google Scholar]
- Bridgham, J.T.; Ortlund, E.A.; Thornton, J.W. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 2009, 461, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Lukeš, J.; Archibald, J.M.; Keeling, P.J.; Doolittle, W.F.; Gray, M.W. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 2011, 63, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.F. Evolutionary biology: A ratchet for protein complexity. Nature 2012, 481, 270–271. [Google Scholar] [CrossRef] [PubMed]
- Mast, F.D.; Barlow, L.D.; Rachubinski, R.A.; Dacks, J.B. Evolutionary mechanisms for establishing eukaryotic cellular complexity. Trends Cell Biol. 2014, 24, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, A. Constructive neutral evolution: Exploring evolutionary theory’s curious disconnec. Biol. Direct 2012, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Literally, “the power of life”. If alive today, Lamarck would surely align himself with those who accept the active agency of organisms. In his time, he vigorously opposed the creationism of Cuvier and could not therefore be interpreted as a creationist.
- Noble, D. Letter from lamarck. Physiol. News 2010, 78, 31. [Google Scholar]
- Noble, D. The Music of Life; OUP: Oxford, UK, 2006. [Google Scholar]
- Pichot, A. Introduction. In Philosophie Zoologique; Flammarion: Paris, France, 1994. [Google Scholar]
- For important modern assessments of Lamarck and Lamarckism see Gissis SB, Jablonka, E. (2015) Transformations of Lamarckism. From subtle fluids to molecular biology. MIT Press).
- Jablonka, E. From replicators to heritably varying phenotypic traits: The extended phenotype revisited. Biol. Philos. 2004, 19, 353–375. [Google Scholar] [CrossRef]
- Noble, D. Evolution beyond neo-darwinism: A new conceptual framework. J. Exp. Biol. 2015, 218, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Noble, D.; Jablonka, E.; Joyner, M.M.; Müller, G.B.; Omholt, S.W. Evolution evolves: Physiology returns to centre stage. J. Physiol. 2014, 592, 2237–2244. [Google Scholar] [CrossRef] [PubMed]
| • Worldwide differences in regional disease frequencies |
| • Low frequency of genetic component of disease as determined with genome wide association studies (GWAS) |
| • Dramatic increases in disease frequencies over past decades |
| • Identical twins with variable and discordant disease frequency |
| • Environmental exposures associated with disease |
| • Regional differences and rapid induction events in evolution |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noble, R.; Noble, D. Was the Watchmaker Blind? Or Was She One-Eyed? Biology 2017, 6, 47. https://doi.org/10.3390/biology6040047
Noble R, Noble D. Was the Watchmaker Blind? Or Was She One-Eyed? Biology. 2017; 6(4):47. https://doi.org/10.3390/biology6040047
Chicago/Turabian StyleNoble, Raymond, and Denis Noble. 2017. "Was the Watchmaker Blind? Or Was She One-Eyed?" Biology 6, no. 4: 47. https://doi.org/10.3390/biology6040047






