Polymorphisms in Interleukin 13 Signaling and Interacting Genes Predict Advanced Fibrosis and Hepatocellular Carcinoma Development in Non-Alcoholic Steatohepatitis
Abstract
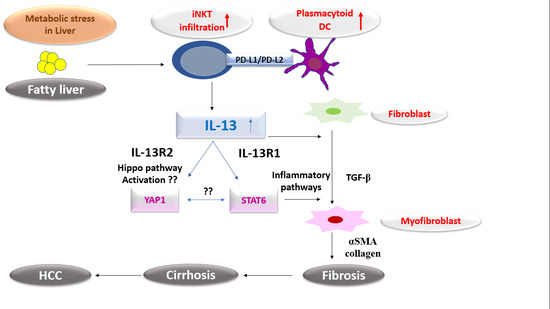
:1. Introduction
2. Subjects and Methods
2.1. Study Population
2.2. Laboratory Analyses
2.3. Genomic DNA Extraction
2.4. Genotyping of Studied Genes
2.5. Serum IL-13 and Serum Insulin Levels
2.6. Statistical Analysis
3. Results
3.1. Study Characteristics of NASH and NASH-HCC Patients
3.2. Association of Adjusted Univariate Significant Parameters with HCC Development in NASH Patients
3.3. IL-13/STAT6 Signaling Axis and YAP1 Are Critical Players for HCC Development in NASH Patients
3.4. Sub-Classifying NASH Patients According to Their Fibrosis Grades
3.5. IL-13/PD-L2 Are Crucial for Fibrosis Progression in NASH Patients
3.6. Comparing Different Fibrosis Grades in NASH with HCC Development
4. Discussion
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2017, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Brunt, E.M.; Wong, V.W.; Nobili, V.; Day, C.P.; Sookoian, S.; Maher, J.J.; Bugianesi, E.; Sirlin, C.B.; Neuschwander-Tetri, B.A.; Rinella, M.E. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers 2015, 1, 15080. [Google Scholar] [CrossRef] [PubMed]
- Starley, B.Q.; Calcagno, C.J.; Harrison, S.A. Nonalcoholic fatty liver disease and hepatocellular carcinoma: A weighty connection. Hepatology 2010, 51, 1820–1832. [Google Scholar] [CrossRef] [PubMed]
- Michelotti, G.A.; Machado, M.V.; Diehl, A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 656–665. [Google Scholar] [CrossRef]
- Arrese, M.; Cabrera, D.; Kalergis, A.M.; Feldstein, A.E. Innate Immunity and Inflammation in NAFLD/NASH. Dig. Dis. Sci. 2016, 61, 1294–1303. [Google Scholar] [CrossRef] [Green Version]
- Hart, K.M.; Fabre, T.; Sciurba, J.C.; Gieseck, R.L., 3rd; Borthwick, L.A.; Vannella, K.M.; Acciani, T.H.; Prado, R.Q.; Thompson, R.; White, S.; et al. Type 2 immunity is protective in metabolic disease but exacerbates NAFLD collaboratively with TGF-beta. Sci. Transl. Med. 2017, 9, eaal3694. [Google Scholar] [CrossRef] [Green Version]
- Gieseck, R.L., 3rd; Wilson, M.S.; Wynn, T.A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 2017, 18, 62–76. [Google Scholar] [CrossRef]
- Nakayama, T.; Hirahara, K.; Onodera, A.; Endo, Y.; Hosokawa, H.; Shinoda, K.; Tumes, D.J.; Okamoto, O. Th2 Cells in Health and Disease. Annu. Rev. Immunol. 2017, 35, 53–84. [Google Scholar] [CrossRef]
- Liu, Y.; Munker, S.; Müllenbach, R.; Weng, H.-L. IL-13 Signaling in Liver Fibrogenesis. Front. Immunol. 2012, 3, 116. [Google Scholar] [CrossRef] [Green Version]
- Weng, H.L.; Liu, Y.; Chen, J.L.; Godoy, P.; Hu, J.H.; Zhou, C.; Stickel, F.; Marx, A.; Bohle, R.M.; Zimmer, V.; et al. The etiology of liver damage imparts cytokines transforming growth factor beta1 or interleukin-13 as driving forces in fibrogenesis. Hepatology 2009, 50, 230–243. [Google Scholar] [CrossRef]
- Shimamura, T.; Fujisawa, T.; Husain, S.R.; Kioi, M.; Nakajima, A.; Puri, R.K. Novel role of IL-13 in fibrosis induced by nonalcoholic steatohepatitis and its amelioration by IL-13R-directed cytotoxin in a rat model. J. Immunol. 2008, 181, 4656–4665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Chierico, F.D.; Sanguinetti, M.; Morelli, D.; Sterbini, F.P.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2018, 69, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Kelly-Welch, A.E.; Hanson, E.M.; Boothby, M.R.; Keegan, A.D. Interleukin-4 and interleukin-13 signaling connections maps. Science 2003, 300, 1527–1528. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Hesse, M.; Sandler, N.G.; Kaviratne, M.; Hoffmann, K.F.; Chiaramonte, M.G.; Reiman, R.; Cheever, A.W.; Sypek, J.P.; Mentink-Kane, M.M. P-selectin suppresses hepatic inflammation and fibrosis in mice by regulating interferon gamma and the IL-13 decoy receptor. Hepatology 2004, 39, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lei, L.; Gu, D.; Liu, H.; Wang, S. CIZ1 is upregulated in hepatocellular carcinoma and promotes the growth and migration of the cancer cells. Tumor Boil. 2015, 37, 4735–4742. [Google Scholar] [CrossRef]
- Xie, M.; Wu, X.-J.; Zhang, J.-J.; He, C.-S. IL-13 receptor α2 is a negative prognostic factor in human lung cancer and stimulates lung cancer growth in mice. Oncotarget 2015, 6, 32902–32913. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, Z.; Caviglia, J.M.; Corey, K.E.; Herfel, T.M.; Cai, B.; Masia, R.; Chung, R.T.; Lefkowitch, J.H.; Schwabeet, R.F.; et al. Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell Metab. 2016, 24, 848–862. [Google Scholar] [CrossRef] [Green Version]
- Kuang, D.M.; Peng, C.; Zhao, Q.; Wu, Y.; Chen, M.S.; Zheng, L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma promote expansion of memory T helper 17 cells. Hepatology 2009, 51, 154–164. [Google Scholar] [CrossRef]
- Tang, S.; Kim, P.S. A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery. Proc. Natl. Acad. Sci. USA 2019, 116, 24500–24506. [Google Scholar] [CrossRef] [Green Version]
- Oldenhove, G.; Boucquey, E.; Taquin, A.; Acolty, V.; Bonetti, L.; Ryffel, B.; Le Bert, M.; Englebert, K.; Boon, L.; Moser, M. PD-1 Is Involved in the Dysregulation of Type 2 Innate Lymphoid Cells in a Murine Model of Obesity. Cell Rep. 2018, 25, 2053–2060.e4. [Google Scholar] [CrossRef] [Green Version]
- Hansel, C.; Erschfeld, S.; Baues, M.; Lammers, T.; Weiskirchen, R.; Trautwein, C.; Kroy, D.C.; Drescher, H.K. The Inhibitory T Cell Receptors PD1 and 2B4 Are Differentially Regulated on CD4 and CD8 T Cells in a Mouse Model of Non-alcoholic Steatohepatitis. Front. Pharmacol. 2019, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; van der Windt, D.J.; Ren, J.; Tsung, A.; Huang, H. The role of neutrophil extracellular traps in nonalcoholic steatohepatitis-associated hepatocellular carcinoma. J. Immunol. 2019, 202 (Suppl. 1), 135.2. [Google Scholar]
- Kim, M.H.; Kim, C.G.; Kim, S.K.; Shin, S.J.; Choe, E.A.; Park, S.H.; Shin, E.C.; Kim, J. YAP-Induced PD-L1 Expression Drives Immune Evasion in BRAFi-Resistant Melanoma. Cancer Immunol. Res. 2018, 6, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.C.; Yang, C.T.; Jablons, D.M.; You, L. The Role of Yes-Associated Protein (YAP) in Regulating Programmed Death-Ligand 1 (PD-L1) in Thoracic Cancer. Biomedicines 2018, 6, 114. [Google Scholar] [CrossRef] [Green Version]
- Janse van Rensburg, H.J.; Azad, T.; Ling, M.; Hao, Y.; Snetsinger, B.; Khanal, P.; Minassian, L.M.; Graham, C.H.; Rauh, M.J.; Yang, X. The Hippo Pathway Component TAZ Promotes Immune Evasion in Human Cancer through PD-L1. Cancer Res. 2018, 78, 1457–1470. [Google Scholar] [CrossRef] [Green Version]
- Shalapour, S.; Lin, X.J.; Bastian, I.N.; Brain, J.; Burt, A.D.; Aksenov, A.A.; Vrbanac, A.F.; Li, W.; Perkins, A.; Matsutaniet, T.; et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 2017, 551, 340–345. [Google Scholar] [CrossRef]
- Jung, H.I.; Jeong, D.; Ji, S.; Ahn, T.S.; Bae, S.H.; Chin, S.; Chung, J.C.; Kim, H.C.; Lee, M.S.; Baek, M.J. Overexpression of PD-L1 and PD-L2 Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma. Cancer Res. Treat. 2017, 49, 246–254. [Google Scholar] [CrossRef] [Green Version]
- Flintoft, L. A SNP for disease prognosis. Nat. Rev. Genet. 2013, 14, 746. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Intraobserver and Interobserver Variations in Liver Biopsy Interpretation in Patients with Chronic Hepatitis, C. The French METAVIR Cooperative Study Group. Hepatology 1994, 20, 15–20.
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Chen, Q.; Zhao, J.; Rafaels, N.; Mathias, P.; Liang, H.; Potee, J.; Campbell, M.; Zhang, B.; Gao, L.; et al. An IL-13 promoter polymorphism associated with liver fibrosis in patients with Schistosoma japonicum. PLoS ONE 2015, 10, e0135360. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Isnard, A.; Kouriba, B.; Cabantous, S.; Dessein, A.; Doumbo, O.; Chevillard, C. A STAT6 gene polymorphism is associated with high infection levels in urinary schistosomiasis. Genes Immun. 2008, 9, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladero, J.M.; Martin, E.G.; Fernandez, C.; Carballo, M.; Devesa, M.J.; Martínez, C.; Suárez, A.; Díaz-Rubio, M.; Agúndez, J.A. Predicting response to therapy in chronic hepatitis C: An approach combining interleukin-28B gene polymorphisms and clinical data. J. Gastroenterol. Hepatol. 2012, 27, 279–285. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Cryer, D. NASH and liver cancer: The new cancer headline. Am. J. Manag. Care 2019, 25, SP334–SP335. [Google Scholar]
- Aleksandrova, K.; Stelmach-Mardas, M.; Schlesinger, S. Obesity and Liver Cancer. Recent Results Cancer Res. 2016, 208, 177–198. [Google Scholar]
- Wynn, T.A.; Vannella, K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 2016, 44, 450–462. [Google Scholar] [CrossRef] [Green Version]
- Matsui, S.; Okabayashi, K.; Tsuruta, M.; Shigeta, K.; Seishima, R.; Ishida, T.; Kondo, T.; Suzuki, Y.; Hasegawa, H.; Shimoda, M.; et al. Interleukin-13 and its signaling pathway is associated with obesity-related colorectal tumorigenesis. Cancer Sci. 2019, 110, 2156–2165. [Google Scholar] [CrossRef]
- Cao, H.; Zhang, J.; Liu, H.; Wan, L.; Zhang, H.; Huang, Q.; Xu, E.; Lai, M. IL-13/STAT6 signaling plays a critical role in the epithelial-mesenchymal transition of colorectal cancer cells. Oncotarget 2016, 7, 61183–61198. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Cai, B.; Li, Y.; Wang, L. P0360: STAT6 rs3024974 might predict worse prognosis in hepatocellular carcinoma patients. J. Hepatol. 2015, 62, S445–S446. [Google Scholar] [CrossRef]
- Lee, Y.L.; Yen, J.J.; Hsu, L.C.; Kuo, N.W.; Su, M.W.; Yang, M.F.; Hsiao, Y.P.; Wang, I.J.; Liu, F.T. Association of STAT6 genetic variants with childhood atopic dermatitis in Taiwanese population. J. Dermatol. Sci. 2015, 79, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Demenais, F.; Margaritte-Jeannin, P.; Barnes, K.C.; Cookson, W.O.C.; Altmüller, J.; Ang, W.; Barr, R.G.; Beaty, T.H.; Becker, A.B.; Beilby, J.; et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat. Genet. 2017, 50, 42–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, M.D.; Gao, P.; Kim, B.E.; Lesley, L.J.; Streib, J.E.; Taylor, P.A.; Zaccaro, D.J.; Boguniewicz, M.; Beck, L.A.; Hanifin, J.M.; et al. The signal transducer and activator of transcription 6 gene (STAT6) increases the propensity of patients with atopic dermatitis toward disseminated viral skin infections. J. Allergy Clin. Immunol. 2011, 128, 1006–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Zhang, Q.; Greten, T.F. Nonalcoholic fatty liver disease promotes hepatocellular carcinoma through direct and indirect effects on hepatocytes. FEBS J. 2017, 285, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mougey, E.B.; Williams, A.; Coyne, A.J.K.; Gutiérrez-Junquera, C.; Fernández-Fernández, S.; Cilleruelo, M.L.; Rayo, A.; Echeverría, L.; Román, E.; Lois, C.G.; et al. CYP2C19 and STAT6 Variants Influence the Outcome of Proton Pump Inhibitor Therapy in Pediatric Eosinophilic Esophagitis. J. Pediatr. Gastroenterol. Nutr. 2019, 69, 581–587. [Google Scholar] [CrossRef]
- Manmadhan, S.; Ehmer, U. Hippo Signaling in the Liver—A Long and Ever-Expanding Story. Front. Cell Dev. Boil. 2019, 7, 33. [Google Scholar] [CrossRef]
- Kodama, T.; Yi, J.; Newberg, J.Y.; Tien, J.C.; Wu, H.; Finegold, M.J.; Kodama, M.; Wei, Z.; Tamura, T.; Takehara, T.; et al. Molecular profiling of nonalcoholic fatty liver disease-associated hepatocellular carcinoma using SB transposon mutagenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E10417–E10426. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Li, W.; Wang, S.; Zhang, P.; Wang, Q.; Xiao, J.; Zhang, C.; Zhang, X.; Xu, X.; Xue, S.; et al. YAP Aggravates Inflammatory Bowel Disease by Regulating M1/M2 Macrophage Polarization and Gut Microbial Homeostasis. Cell Rep. 2019, 27, 1176–1189.e5. [Google Scholar] [CrossRef] [Green Version]
- Zhubanchaliyev, A.; Temirbekuly, A.; Kongrtay, K.; Wanshura, L.C.; Kunz, J. Targeting Mechanotransduction at the Transcriptional Level: YAP and BRD4 Are Novel Therapeutic Targets for the Reversal of Liver Fibrosis. Front. Pharmacol. 2016, 7, 462. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.J.; Ju, Q.; Li, G.C. Tumor markers for hepatocellular carcinoma. Mol. Clin. Oncol. 2013, 1, 593–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.L.; Patman, G.L.; Leathart, J.B.; Piguet, A.C.; Burt, A.D.; Dufour, J.F.; Day, C.P.; Daly, A.K.; Reeves, H.L.; Anstee, Q.M.; et al. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J. Hepatol. 2014, 61, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Tokushige, K.; Hashimoto, E.; Horie, Y.; Taniai, M.; Higuchi, S. Hepatocellular carcinoma in Japanese patients with nonalcoholic fatty liver disease, alcoholic liver disease, and chronic liver disease of unknown etiology: Report of the nationwide survey. J. Gastroenterol. 2011, 46, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Das, K.; Mukherjee, P.S.; Ghosh, A.; Ghosh, S.; Mridha, A.R.; Dhibar, T.; Bhattacharya, B.; Bhattacharya, D.; Manna, B.; et al. Nonobese population in a developing country has a high prevalence of nonalcoholic fatty liver and significant liver disease. Hepatology 2010, 51, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Dondeti, M.F.; El-Maadawy, E.A.; Talaat, R.M. Hepatitis-related hepatocellular carcinoma: Insights into cytokine gene polymorphisms. World J. Gastroenterol. 2016, 22, 6800–6816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maricic, I.; Marrero, I.; Eguchi, A.; Nakamura, R.; Johnson, C.D.; Dasgupta, S.; Hernandez, C.D.; Nguyen, P.S.; Swafford, A.D.; Knight, R.; et al. Differential Activation of Hepatic Invariant NKT Cell Subsets Plays a Key Role in Progression of Nonalcoholic Steatohepatitis. J. Immunol. 2018, 201, 3017–3035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegde, S.; Lockridge, J.L.; Becker, Y.A.; Ma, S.; Kenney, S.C.; Gumperz, J.E. Human NKT cells direct the differentiation of myeloid APCs that regulate T cell responses via expression of programmed cell death ligands. J. Autoimmun. 2011, 37, 28–38. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, S.; Surette, F.A.; Hahn, Y.S. The Immune Landscape in Nonalcoholic Steatohepatitis. Immune Netw. 2016, 16, 147–158. [Google Scholar] [CrossRef] [Green Version]

| Characteristics | NASH (n = 79) | NASH-HCC (n = 55) | p-Value | Statistics (Univariate) OR (95%CI) |
|---|---|---|---|---|
| Age mean ± SD (years) | 61.2 ± 5.9 | 63.9 ± 6.4 | 0.017 | 1.073 (1.012–1.138) |
| BMI, median (range), kg/m2 | 29 (4.3) | 28.3 (7.3) | - | |
| a AST median (range), U/L | 53 (35) | 67 (18) | - | |
| b ALT median (range), U/L | 48 (21) | 44 (11) | - | |
| Albumin median (range), g/dL | 3.7 (0.5) | 3.3 (0.7) | - | |
| Bilirubin median (range), mg/dL | 1.5 (0.8) | 1.3 (0.7) | - | |
| Hemoglobin median (range), g/dL | 13.4 (2.7) | 12 (1.7) | - | |
| Platelets median (range), 109/L | 178 (84) | 129 (97) | - | |
| c TLC median (range), 109/µL | 6.5 (2.7) | 6 (1.4) | - | |
| d TAG median (range), mg/dL | 158 (60) | 160 (47) | - | |
| Glucose median (range), mg/dL | 101 (18) | 106 (22) | - | |
| Insulin median (range), µU/mL | 11.6 (3.3) | 12 (3.7) | - | |
| e HOMA-IR median (range) | 3 (0.9) | 3.2 (1) | - | |
| f AFP median (range), ng/mL | 9.5 (5) | 407.7 (570) | <0.001 | 7.641 (3.6–16.18) |
| IL-13 mean ± SD, (ng/L) | 4.9 ± 1.7 | 7.85 ± 4.3 | <0.001 | 1.616 (1.3–2.008) |
| Fibrosis stage, [n(%)] | <0.001 | |||
| F1–F2 | 41 (51.8) | - | - | |
| F3–F4 | 38 (48.1) | 5 (9) | - | |
| Cirrhosis | - | 50 (90.9) | - |
| Genotype/Allele | NASH (n = 79) | NASH-HCC (n = 55) | Chi2 p-value | OR (95% CI) | |
|---|---|---|---|---|---|
| IL-13 rs20541 [n(%)] | Chi2 = 12.12, p = 0.002 | ||||
| A/A | 3 (3.7) | 12 (21.8) | |||
| A/G | 30 (37.9) | 12 (21.8) | |||
| G/G | 46 (58.2) | 31 (56.3) | chi2 = 10.5 0.004 | 7.07 (1.89–26.44) | |
| IL-13R1 rs2248841 [n(%)] | Chi2 = 1.854, p = 0.396 | ||||
| C/C | 6 (7.5) | 8 (14.5) | |||
| T/C | 18 (22.7) | 10 (18.1) | |||
| T/T | 55 (69.6) | 37 (67.2) | |||
| IL-13R2 rs5946040 [n(%)] | Chi2 = 6.09, p = 0.048 | ||||
| G/G | 9 (11.3) | 14 (25.4) | |||
| G/T | 6 (7.5) | 1 (1.8) | |||
| T/T | 64 (81) | 40 (72.7) | chi2 = 4.5 0.038 | 2.656 (1.056–6.677) | |
| STAT6 rs167769 [n(%)] | Chi2 = 24.4, p < 0.001 | ||||
| C/C | 70 (88.6) | 29 (52.7) | chi2 = 21.6 <0.001 | 0.124 (0.052–0.297) | |
| C/T | 6 (7.5) | 24 (43.6) | |||
| T/T | 3 (3.7) | 2 (3.6) | |||
| YAP1 rs11225163 [n(%)] | Chi2 = 6.14, p = 0.046 | ||||
| C/C | 39 (49.3) | 20 (36.3) | |||
| C/T | 34 (43) | 23 (41.8) | |||
| T/T | 6 (7.5) | 12 (21.8) | |||
| C allele carriers/C allele non carriers | 73/6 | 43/12 | chi2 = 5.56 0.022 | 0.295 (0.103–0.842) | |
| PD-L1 rs2282055 [n(%)] | Chi2 = 6.273, p = 0.043 | ||||
| G/G | 5 (6.3) | 11 (2) | |||
| G/T | 12 (15.1) | 5 (9) | |||
| T/T | 62 (78) | 39 (70.9) | chi2 = 5.76 0.022 | 3.7 (1.206–11.352) | |
| PD-L2 rs7854413 [n(%)] | Chi2 = 1.99, p = 0.37 | ||||
| C/C | 5 (6.3) | 6 (10.9) | |||
| T/C | 23 (29.1) | 11 (20) | |||
| T/T | 51 (64.5) | 38 (69) | |||
| Parameters | Univariate Regression Analysis | Multivariate Regression Analysis | ||
|---|---|---|---|---|
| Adjusted OR (95%CI) | p-value | Adjusted OR (95%CI) | p-value | |
| * AFP ng/mL | 7.854 (3.58–17.2) | <0.001 | 19.6 (4.36–88.85) | <0.001 |
| IL-13 ng/L | 1.727 (1.368–2.18) | <0.001 | 1.9 (1.211–2.99) | 0.005 |
| IL-13 rs20541, G/G genotype | 6.172 (1.622–23.47) | 0.008 | - | 0.491 |
| IL-13R2 rs5946040, T/T genotype | 3.5 (1.301–9.445) | 0.013 | - | 0.832 |
| STAT6 rs167769, C/C genotype | 0.092 (0.035–0.244) | <0.001 | 0.015 (0.002–0.123) | <0.001 |
| YAP1 rs11225163, C allele carriers/C allele non carriers | 0.213 (0.07–0.647) | 0.006 | 0.047 (0.006–0.386) | 0.004 |
| PD-L1 rs2282055 T/T genotype | 4.015 (1.272–12.669) | 0.018 | - | 0.598 |
| Genotype/Allele | NASH F1–F2 (n = 40) | NASH F3–F4 (n = 39) | Chi2 p-value | OR (95% CI) | |
|---|---|---|---|---|---|
| IL-13 rs20541 [n(%)] | Chi2 = 9.963, p = 0.007 | ||||
| A/A | 3 (7.5) | - | |||
| A/G | 9 (22.5) | 21 (53.8) | |||
| G/G | 28 (70) | 18 (46.1) | |||
| A allele carriers/A allele non carriers | 12/40 | 21/39 | chi2 = 4.617 0.034 | 2.722 (1.081–6.6858) | |
| IL-13R1 rs2248841 [n(%)] | Chi2 = 2.879, p = 0.237 | ||||
| C/C | 3 (7.5) | 3 (7.7) | |||
| T/C | 6 (15) | 12 (30.7) | |||
| T/T | 31 (77.5) | 24 (61.5) | |||
| IL-13R2 rs5946040 [n(%)] | Chi2 = 7.89, p = 0.018 | ||||
| G/G | 6 (15) | 3 (7.6) | |||
| G/T | 6 (15) | ||||
| T/T | 28 (70) | 36 (92.3) | |||
| G allele carriers/G allele non carriers | 12/40 | 3/39 | chi2 = 6.388 0.018 | 0.194 (0.05–0.756) | |
| STAT6 rs167769 [n(%)] | Chi2 = 1.045, p = 0.593 | ||||
| C/C | 34 (85) | 36 (92) | |||
| C/T | 4 (10) | 2 (5.1) | |||
| T/T | 2 (5) | 1 (2.5) | |||
| YAP1 rs11225163 [n(%)] | Chi2 = 6.69, p = 0.127 | ||||
| C/C | 21 (52.5) | 18 (46.1) | |||
| C/T | 19 (47.5) | 15 (38.4) | |||
| T/T | 6 (15.3) | ||||
| PD-L1 rs2282055 [n(%)] | Chi2 = 0.252, p = 0.882 | ||||
| G/G | 2 (5) | 3 (7.6) | |||
| G/T | 6 (15) | 6 (15.3) | |||
| T/T | 32 (80) | 30 (37.9) | |||
| PD-L2 rs7854413 [n(%)] | Chi2 = 12.923, p = 0.002 | ||||
| C/C | 5 (12.8) | ||||
| T/C | 7 (17.5) | 16 (41) | |||
| T/T | 33 (82.5) | 18 (46.1) | |||
| C allele carriers/C allele non carriers | 7/40 | 21/39 | chi2 = 7.495 0.001 | 5.5 (1.963–115.411) | |
| Parameters | Univariate Regression Analysis | Multivariate Regression Analysis | ||
|---|---|---|---|---|
| OR (95%CI) | p-value | OR (95%CI) | p-value | |
| IL-13 ng/L | 1.496 (1.111–2.014) | 0.008 | 1.432 (1.022–2.008) | 0.037 |
| IL-13 rs20541, A allele carriers/A allele non carriers | 2.722 (1.081–6.685) | 0.034 | - | 0.118 |
| IL-13R2 rs5946040, G allele carriers/G allele non carriers | 0.194 (0.05–0.756) | 0.018 | - | 0.072 |
| PD-L2 rs7854413, C allele carriers/C allele non carriers | 5.5 (1.963–115.411) | 0.001 | 3.797 (1.216–11.875) | 0.022 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Derany, M.O. Polymorphisms in Interleukin 13 Signaling and Interacting Genes Predict Advanced Fibrosis and Hepatocellular Carcinoma Development in Non-Alcoholic Steatohepatitis. Biology 2020, 9, 75. https://doi.org/10.3390/biology9040075
El-Derany MO. Polymorphisms in Interleukin 13 Signaling and Interacting Genes Predict Advanced Fibrosis and Hepatocellular Carcinoma Development in Non-Alcoholic Steatohepatitis. Biology. 2020; 9(4):75. https://doi.org/10.3390/biology9040075
Chicago/Turabian StyleEl-Derany, Marwa O. 2020. "Polymorphisms in Interleukin 13 Signaling and Interacting Genes Predict Advanced Fibrosis and Hepatocellular Carcinoma Development in Non-Alcoholic Steatohepatitis" Biology 9, no. 4: 75. https://doi.org/10.3390/biology9040075