Molecular Analysis of Membrane Targeting by the C2 Domain of the E3 Ubiquitin Ligase Smurf1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Smurf1 Full-Length Site-Directed Mutagenesis
2.3. Protein Purification
2.4. Liposome Binding Assays
2.5. Snooper Lipid Detection
2.6. SPR Spectroscopy
2.7. Cloning and Plasmids
2.8. Cell Culture and Transfection
2.9. Microscopy and Live Cell Imaging
2.10. Quantitative Image Analysis
2.11. Live Cell Ubiquitination Assay
2.12. Docking and Molecular Dynamics Simulations
3. Results
3.1. The Smurf1 C2 Domain Binds PIPs and Phosphatidylserine In Vitro
3.2. Smurf1 C2 Localizes to the Plasma Membrane and Intracellular Anionic Membranes
3.3. Analysis of C2 Mutants Indicates the Loop Region is Necessary for Membrane Localization
3.4. Smurf1 C2 Localization to the Plasma Membrane Is PIP Dependent
3.5. RFP-Ubiquitin Localization is Dependent on the Ligase Activity of Smurf1 at the Membrane
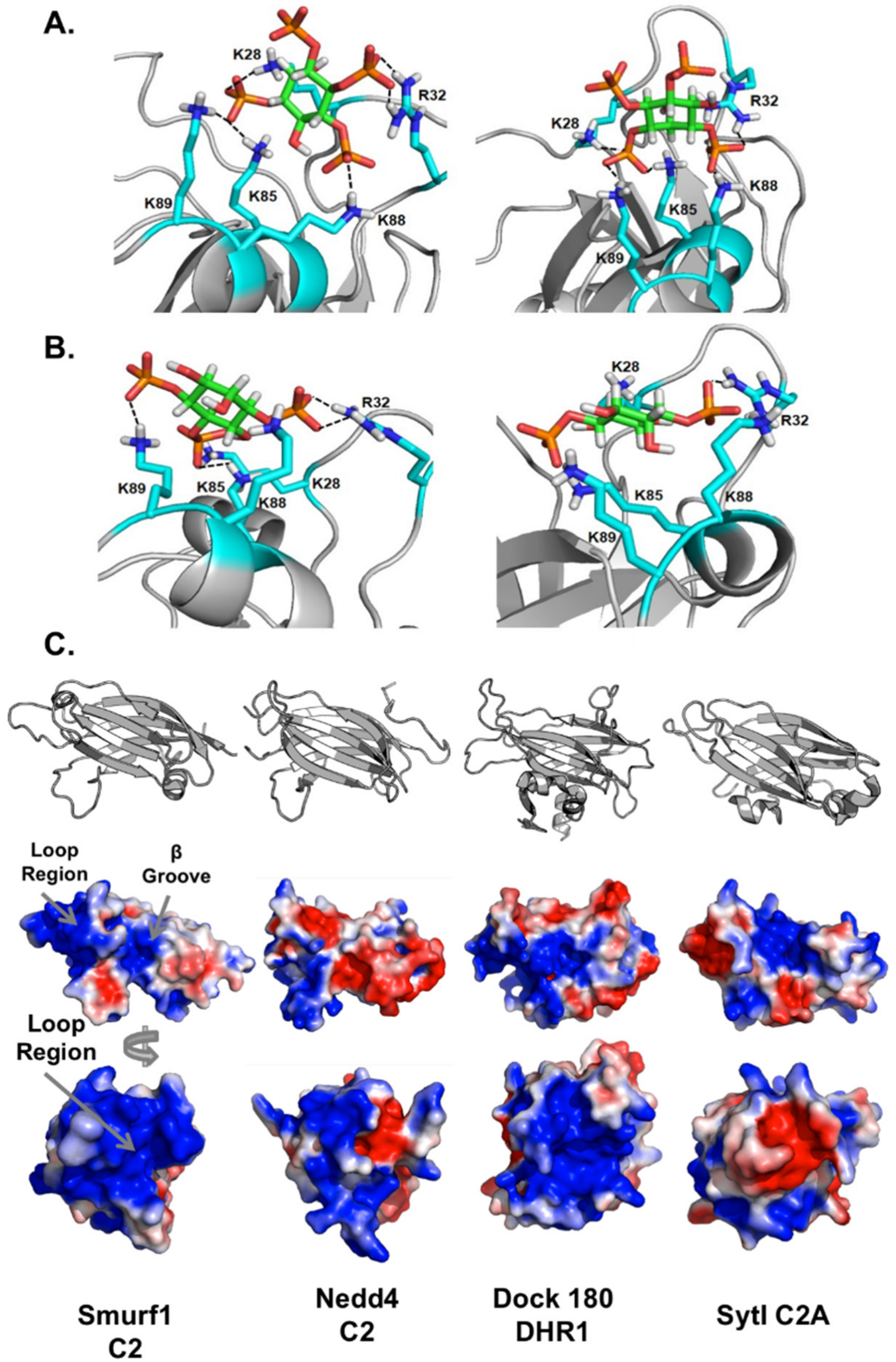
3.6. Computational Analysis of PIP Binding Reveals Two Potential Binding Orientations
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suzuki, C.; Murakami, G.; Fukuchi, M.; Shimanuki, T.; Shikauchi, Y.; Imamura, T.; Miyazono, K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 2002, 277, 39919–39925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.R.; Zhang, Y.; Ozdamar, B.; Ogunjimi, A.A.; Alexandrova, E.; Thomsen, G.H.; Wrana, J.L. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 2003, 302, 1775–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, S.X.; Hussain, Z.J.; Zhang, Y.E. Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein-2-induced osteoblast conversion by targeting Smad5 for degradation. J. Biol. Chem. 2003, 278, 39029–39036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, K.; Fujita, T.; Ozaki, T.; Kato, C.; Kurose, Y.; Sakamoto, M.; Kato, S.; Goto, T.; Itoyama, Y.; Aoki, M.; et al. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J. Biol. Chem. 2004, 279, 11327–11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuda, J.; Nakao, M.; Kawaoka, Y.; Shida, H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 2003, 77, 9987–9992. [Google Scholar] [CrossRef] [Green Version]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Asano, Y.; Ihn, H.; Yamane, K.; Kubo, M.; Tamaki, K. Impaired Smad7-Smurf-mediated negative regulation of TGF-β signaling in scleroderma fibroblasts. J. Clin. Investig. 2004, 113, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Bonni, S.; Wang, H.R.; Causing, C.G.; Kavsak, P.; Stroschein, S.L.; Luo, K.; Wrana, J.L. TGF-β induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol. 2001, 3, 587–595. [Google Scholar] [CrossRef]
- Ebisawa, T.; Fukuchi, M.; Murakami, G.; Chiba, T.; Tanaka, K.; Imamura, T.; Miyazono, K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001, 276, 12477–12480. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.; Li, P.; Zhang, M.; Xing, G.; Li, X.; Zhou, W.; Bartlam, M.; Zhang, L.; Rao, Z.; He, F. Pivotal role of the C2 domain of the Smurf1 ubiquitin ligase in substrate selection. J. Biol. Chem. 2011, 286, 16861–16870. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Lu, K.; Wang, J.; An, L.; Yang, G.; Chen, H.; Cui, Y.; Yin, X.; Xie, P.; Xing, G.; et al. Ubiquitin ligase Smurf1 targets TRAF family proteins for ubiquitination and degradation. Mol. Cell. Chem. 2010, 338, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jin, C.; Tang, Y.; Tang, L.Y.; Zhang, Y.E. Ubiquitination of tumor necrosis factor receptor-associated factor 4 (TRAF4) by Smad ubiquitination regulatory factor 1 (Smurf1) regulates motility of breast epithelial and cancer cells. J. Biol. Chem. 2013, 288, 21784–21792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narimatsu, M.; Bose, R.; Pye, M.; Zhang, L.; Miller, B.; Ching, P.; Sakuma, R.; Luga, V.; Roncari, L.; Attisano, L.; et al. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell 2009, 137, 295–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, P.L.; Lu, H.; Shelly, M.; Geo, H.F.; Poo, M.M. Phosphorylation of E3 Ligase Smurf1 Switches Its Substrate Preference in Support of Axon Development. Neuron 2011, 69, 231–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, M.; Ying, S.X.; Zhang, G.M.; Li, C.; Cheng, S.Y.; Deng, C.X.; Zhang, Y.E. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 2005, 121, 101–113. [Google Scholar] [CrossRef] [Green Version]
- Orvedahl, A.; Sumpter, R.; Xiao, G.H.; Ng, A.; Zou, Z.J.; Tang, Y.; Narimatsu, M.; Gilpin, C.; Sun, Q.H.; Roth, M.; et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 2011, 480, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Yoon, Y.; Tong, J.; Lee, P.J.; Albanese, A.; Bhardwaj, N.; Kallberg, M.; Digman, M.A.; Lu, H.; Gratton, E.; Shin, Y.K.; et al. Molecular basis of the potent membrane-remodeling activity of the epsin 1 N-terminal homology domain. J. Biol. Chem. 2010, 285, 531–540. [Google Scholar] [CrossRef] [Green Version]
- Cho, W.; Stahelin, R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 119–151. [Google Scholar] [CrossRef]
- Cho, W.; Stahelin, R.V. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 2006, 1761, 838–849. [Google Scholar] [CrossRef]
- Stahelin, R.V.; Rafter, J.D.; Das, S.; Cho, W. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J. Biol. Chem. 2003, 278, 12452–12460. [Google Scholar] [CrossRef] [Green Version]
- Corbalan-Garcia, S.; Gomez-Fernandez, J.C. The C2 domains of classical and novel PKCs as versatile decoders of membrane signals. Biofactors 2010, 36, 1–7. [Google Scholar] [PubMed]
- Guillen, J.; Ferrer-Orta, C.; Buxaderas, M.; Perez-Sanchez, D.; Guerrero-Valero, M.; Luengo-Gil, G.; Pous, J.; Guerra, P.; Gomez-Fernandez, J.C.; Verdaguer, N.; et al. Structural insights into the Ca2+ and PI(4,5)P-2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1. Proc. Natl Acad. Sci. USA 2013, 110, 20503–20508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duning, K.; Wennmann, D.O.; Bokemeyer, A.; Reissner, C.; Wersching, H.; Thomas, C.; Buschert, J.; Guske, K.; Franzke, V.; Floel, A.; et al. Common exonic missense variants in the C2 domain of the human KIBRA protein modify lipid binding and cognitive performance. Transl. Psych. 2013, 3, e272. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.R.V.; Fischer, M.J.; Anderson, K.E.; Holdich, J.; Koteci, A.; Balla, T.; Irvine, R.F. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 2012, 337, 727–730. [Google Scholar] [CrossRef] [Green Version]
- Schrodinger Protein Preparation Wizard, 2014-1; Schrodinger LLC: New York, NY, USA, 2014; Available online: www.schrodinger.com/protein-preparation-wizard (accessed on 31 December 2019).
- Glide Schrodinger, 2014-1; Schrodinger LLC: New York, NY, USA, 2014; Available online: www.schrodinger.com/glide (accessed on 31 December 2019).
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, I.T.E.; Darden, T.A. AMBER, v14; University of California: San Francisco, CA, USA, 2014; Available online: www.ambermd.org (accessed on 31 December 2019).
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Pastor, R.W.; Brooks, B.R.; Szabo, A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 1988, 65, 1409–1419. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Scott, J.L.; Musselman, C.A.; Adu-Gyamfi, E.; Kutateladze, T.G.; Stahelin, R.V. Emerging methodologies to investigate lipid-protein interactions. Integr. Biol. 2012, 4, 247–258. [Google Scholar] [CrossRef] [Green Version]
- Stahelin, R.V.; Scott, J.L.; Frick, C.T. Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chem. Phys. Lipids 2014, 182, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Stahelin, R.V. Lipid binding domains: More than simple lipid effectors. J. Lipid Res. 2009, 50, S299–S304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manna, D.; Bhardwaj, N.; Vora, M.S.; Stahelin, R.V.; Lu, H.; Cho, W. Differential roles of phosphatidylserine, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in plasma membrane targeting of C2 domains. Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKCα C2 domain. J. Biol. Chem. 2008, 283, 26047–26058. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.H.; Murray, D.; Leslie, C.C.; Falke, J.J. Specific translocation of protein kinase Cα to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol. Biol. Cell 2006, 17, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, S.; Shirai, T.; Kiyokawa, E.; Mochizuki, N.; Matsuda, M.; Fukui, Y. Membrane recruitment of DOCK180 by binding to Ptdlns(3,4,5)P3. Biochem. J. 2001, 354, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Premkumar, L.; Bobkov, A.A.; Patel, M.; Jaroszewski, L.; Bankston, L.A.; Stec, B.; Vuori, K.; Cote, J.F.; Liddington, R.C. Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs). J. Biol. Chem. 2010, 285, 13211–13222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Matteis, M.A.; Godi, A.; Di Campli, A.; Konstantakopoulos, A.; Di Tullio, G.; Alessi, D.R.; Kular, G.S.; Daniele, T.; Marra, P.; Lucocq, J.M. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004, 6, 393–404. [Google Scholar]
- Fairn, G.D.; Hermansson, M.; Somerharju, P.; Grinstein, S. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat. Cell Biol. 2011, 13, 1424–1498. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Ohara, O.; Ando, A.; Nagase, T. Smurf1 directly targets hPEM-2, a GEF for Cdc42, via a novel combination of protein interaction modules in the ubiquitin-proteasome pathway. Biol. Chem. 2008, 389, 405–413. [Google Scholar] [CrossRef]
- Li, H.; Xiao, N.; Wang, Y.; Wang, R.; Chen, Y.; Pan, W.; Liu, D.; Li, S.; Sun, J.; Zhang, K.; et al. Smurf1 regulates lung cancer cell growth and migration through interaction with and ubiquitination of PIPKIγ. Oncogene 2017, 36, 5668–5680. [Google Scholar] [CrossRef]
- Fei, C.; He, X.; Xie, S.; Miao, H.; Zhou, Z.; Li, L. Smurf1-mediated axin ubiquitination requires Smurf1 C2 domain and is cell cycle-dependent. J. Biol. Chem. 2014, 289, 14170–14177. [Google Scholar] [CrossRef] [Green Version]
- Dall’Armi, C.; Devereaux, K.A.; Di Paolo, G. The role of lipids in the control of autophagy. Curr. Biol. 2013, 23, R33–R45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, T.; Heit, B.; Dubuisson, J.F.; Fairn, G.D.; Chiu, B.; Inman, R.; Kapus, A.; Swanson, M.; Grinstein, S. Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. J. Cell Biol. 2009, 185, 917–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Jia, Y.; Zhang, Y.; Ma, F.; Zhu, Y.; Hong, X.; Zhou, Q.; He, R.; Zhang, H.; Jin, J.; et al. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy 2020, 16, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.H.; Nair, V.R.; Scharn, C.R.; Xavier, R.J.; Torrealba, J.R.; Shiloh, M.U.; Levine, B. The ubiquitin ligase Smurf1 functions in selective autophagy of mycobacterium tuberculosis and anti-tuberculosis host defense. Cell Host Microbe 2017, 21, 59–72. [Google Scholar] [CrossRef] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scott, J.L.; Frick, C.T.; Johnson, K.A.; Liu, H.; Yong, S.S.; Varney, A.G.; Wiest, O.; Stahelin, R.V. Molecular Analysis of Membrane Targeting by the C2 Domain of the E3 Ubiquitin Ligase Smurf1. Biomolecules 2020, 10, 229. https://doi.org/10.3390/biom10020229
Scott JL, Frick CT, Johnson KA, Liu H, Yong SS, Varney AG, Wiest O, Stahelin RV. Molecular Analysis of Membrane Targeting by the C2 Domain of the E3 Ubiquitin Ligase Smurf1. Biomolecules. 2020; 10(2):229. https://doi.org/10.3390/biom10020229
Chicago/Turabian StyleScott, Jordan L., Cary T. Frick, Kristen A. Johnson, Haining Liu, Sylvia S. Yong, Allyson G. Varney, Olaf Wiest, and Robert V. Stahelin. 2020. "Molecular Analysis of Membrane Targeting by the C2 Domain of the E3 Ubiquitin Ligase Smurf1" Biomolecules 10, no. 2: 229. https://doi.org/10.3390/biom10020229





