FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin and Trypsin under Neutral Salts
Abstract
:1. Introduction
- the adsorption of ions on the protein surface destabilizes the protein structure,
- the ions are adsorbed if they have an affinity for the protein surface,
- the affinity for the protein surface can be predicted using LMWA.
2. Materials and Methods
3. Results and Discussion
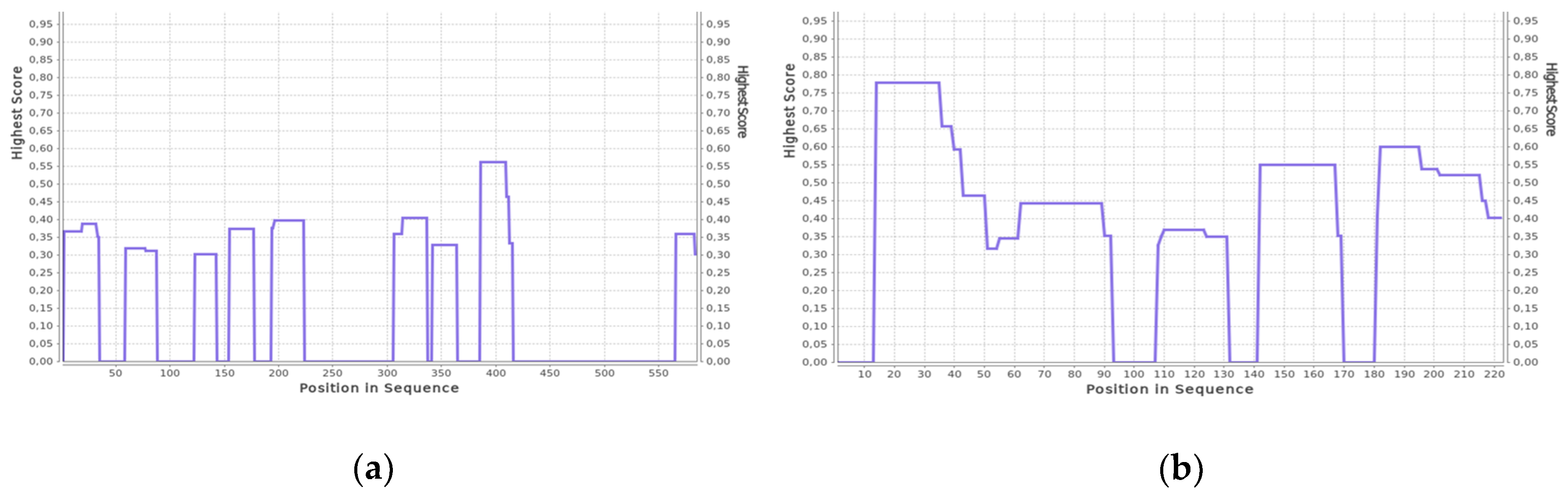
3.1. Spectral Studies of Native Protein Structures
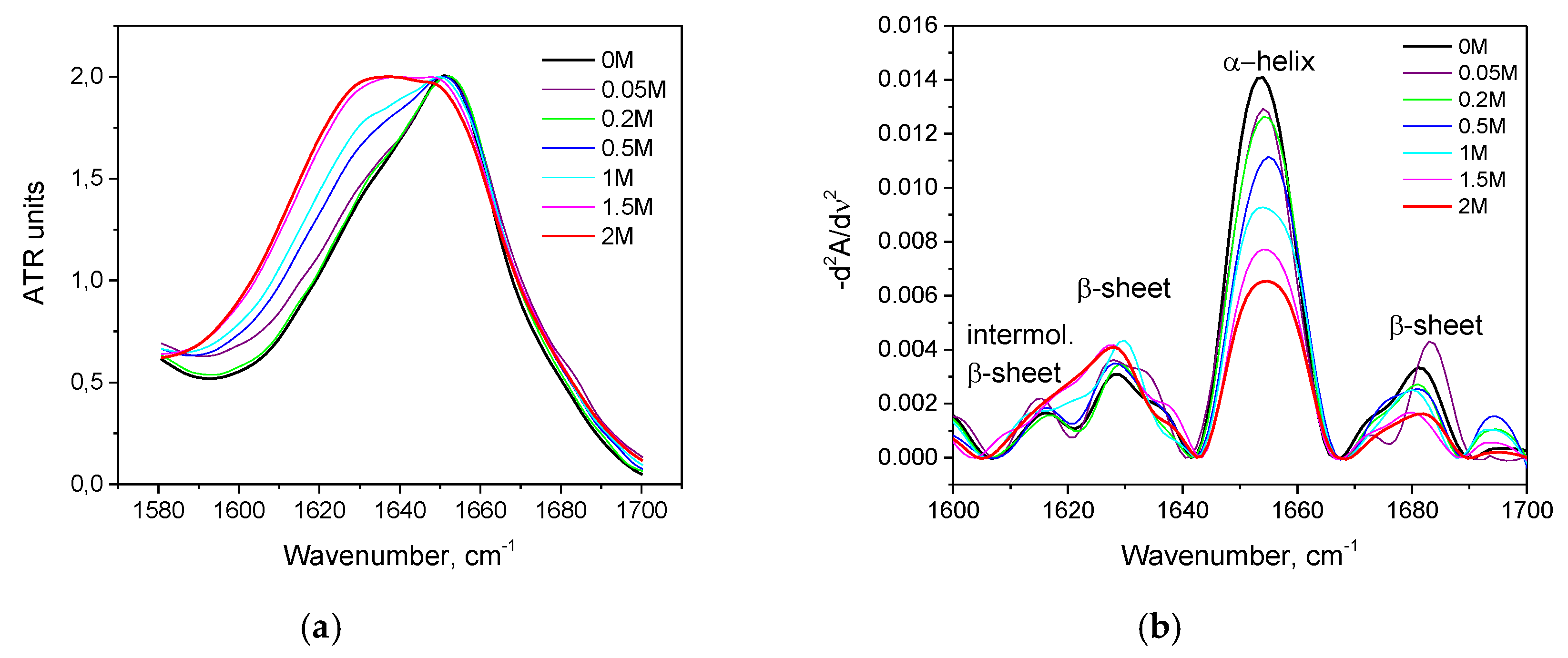
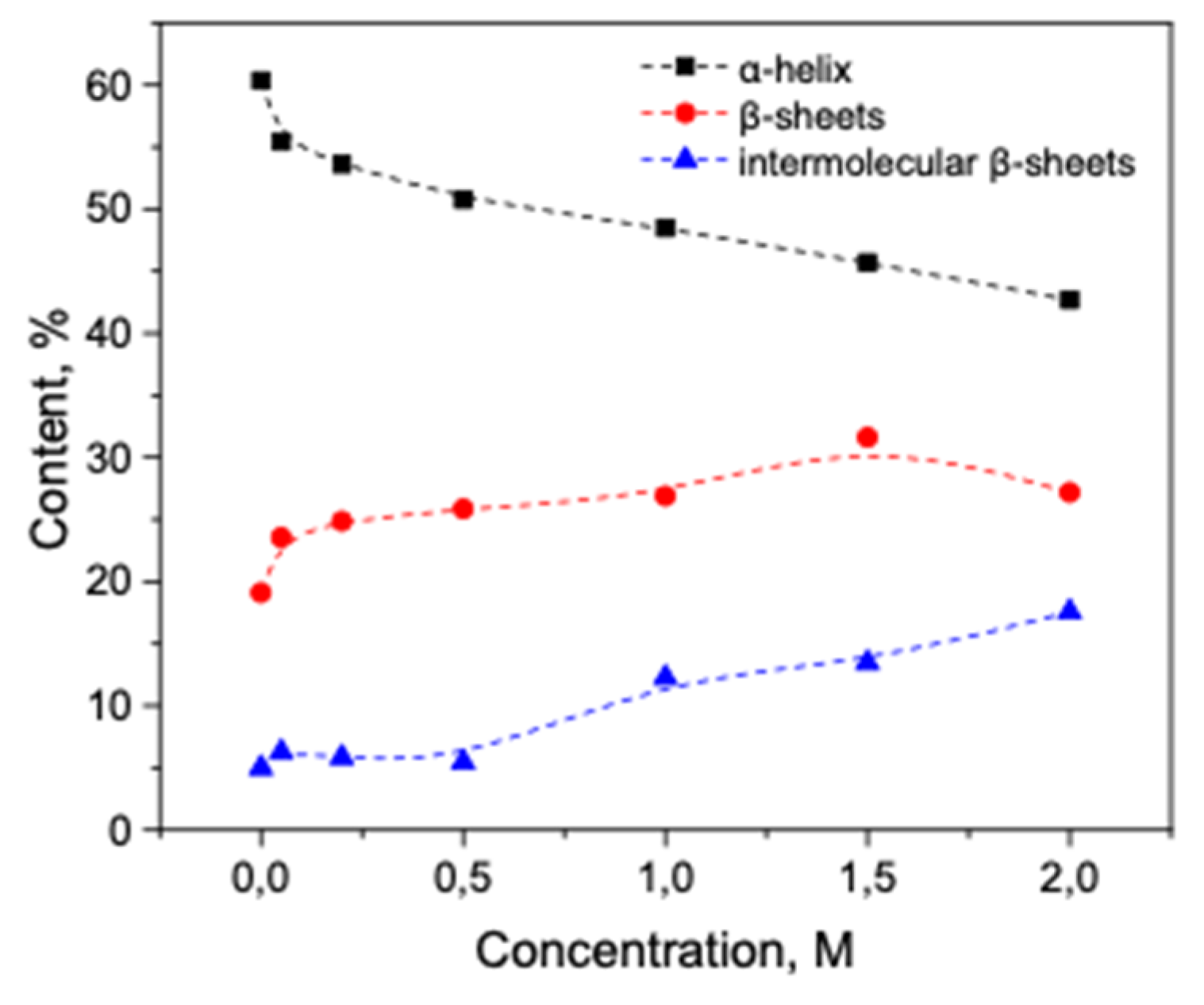
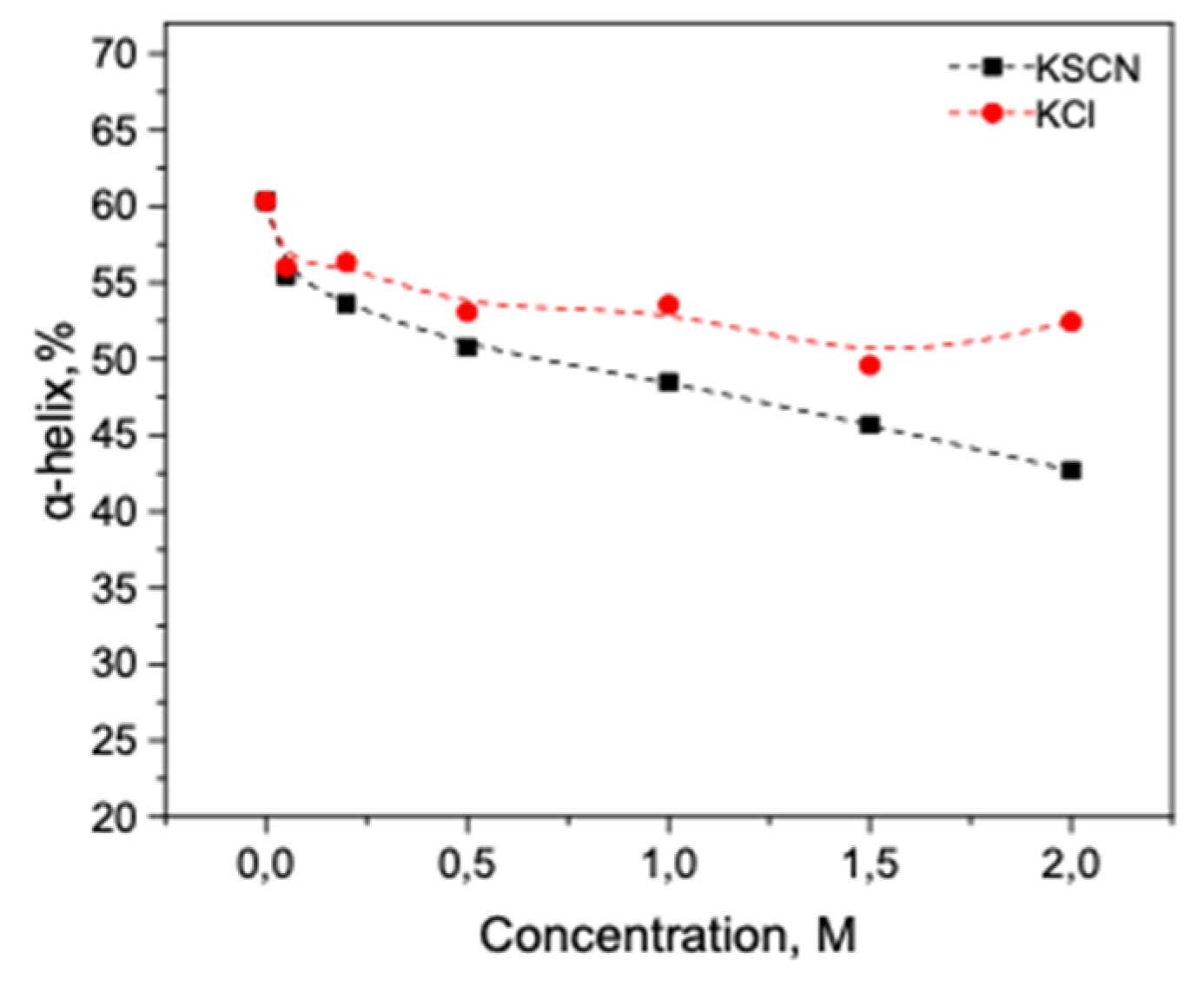
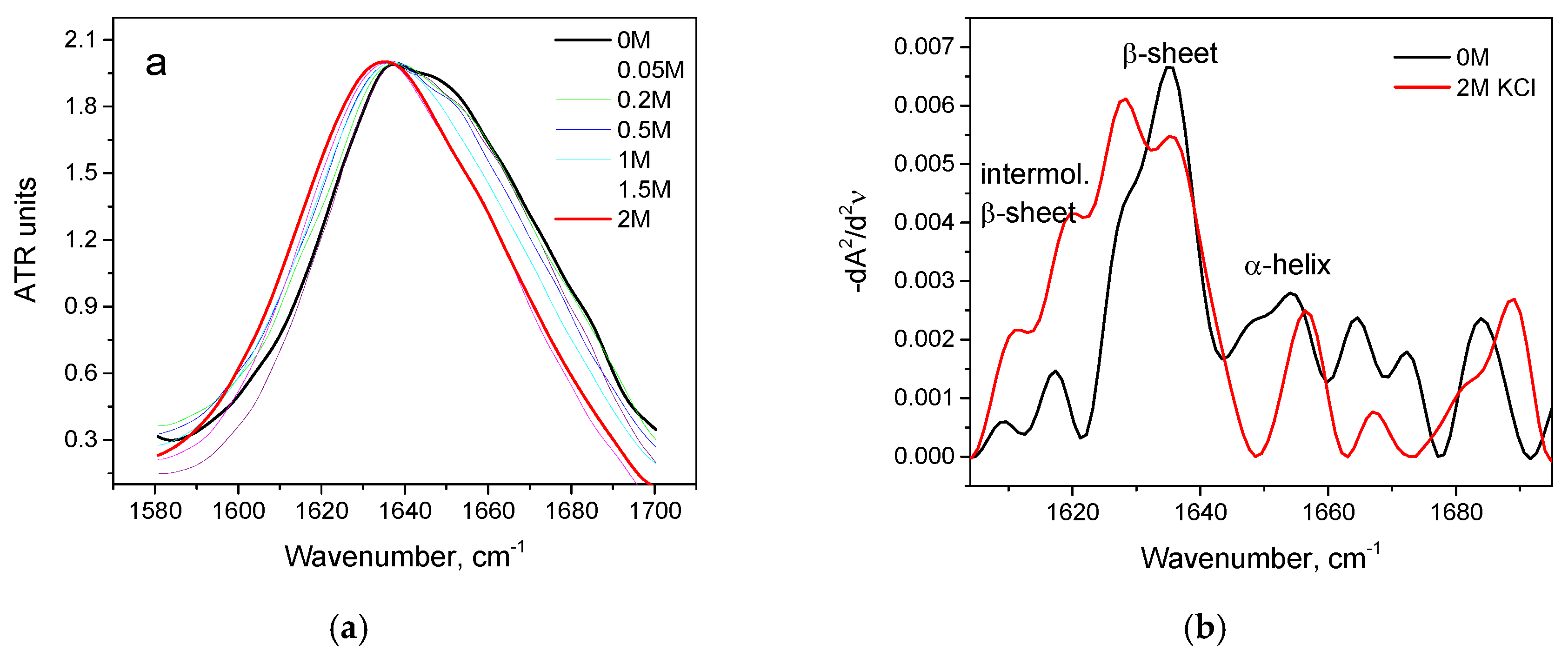
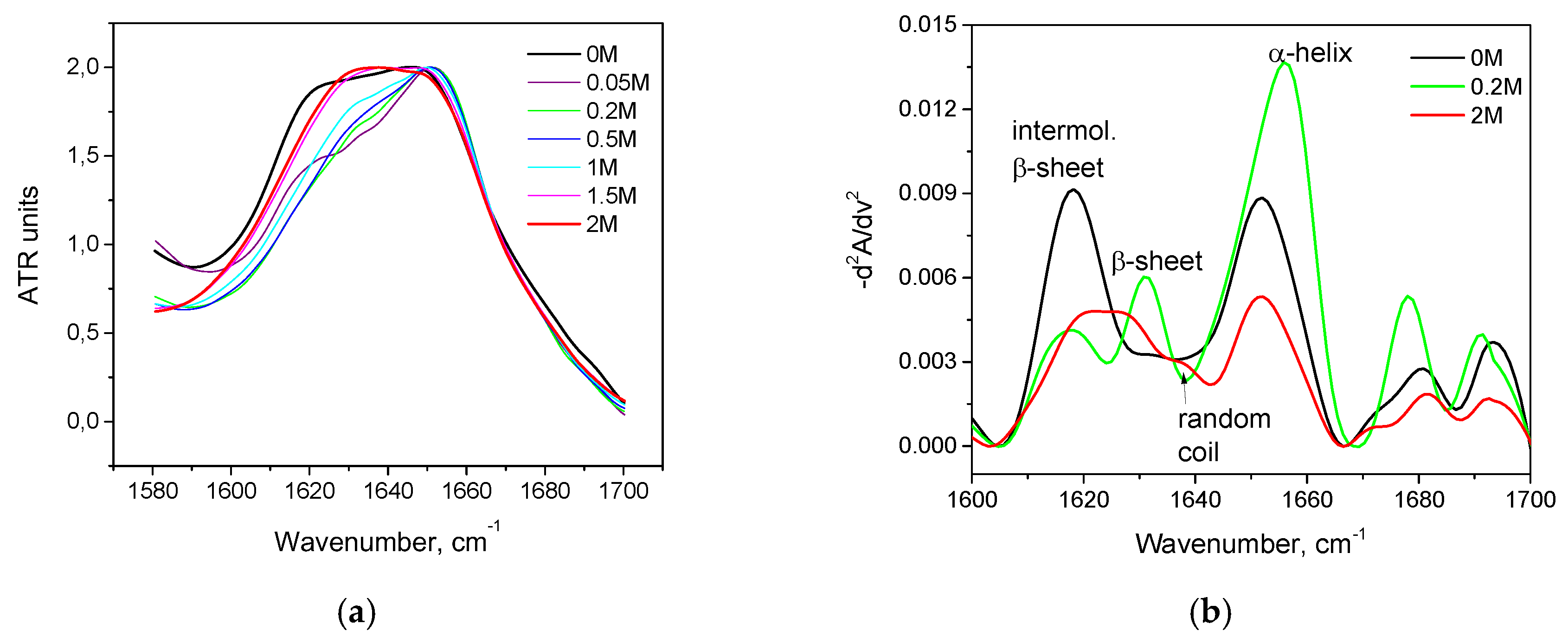
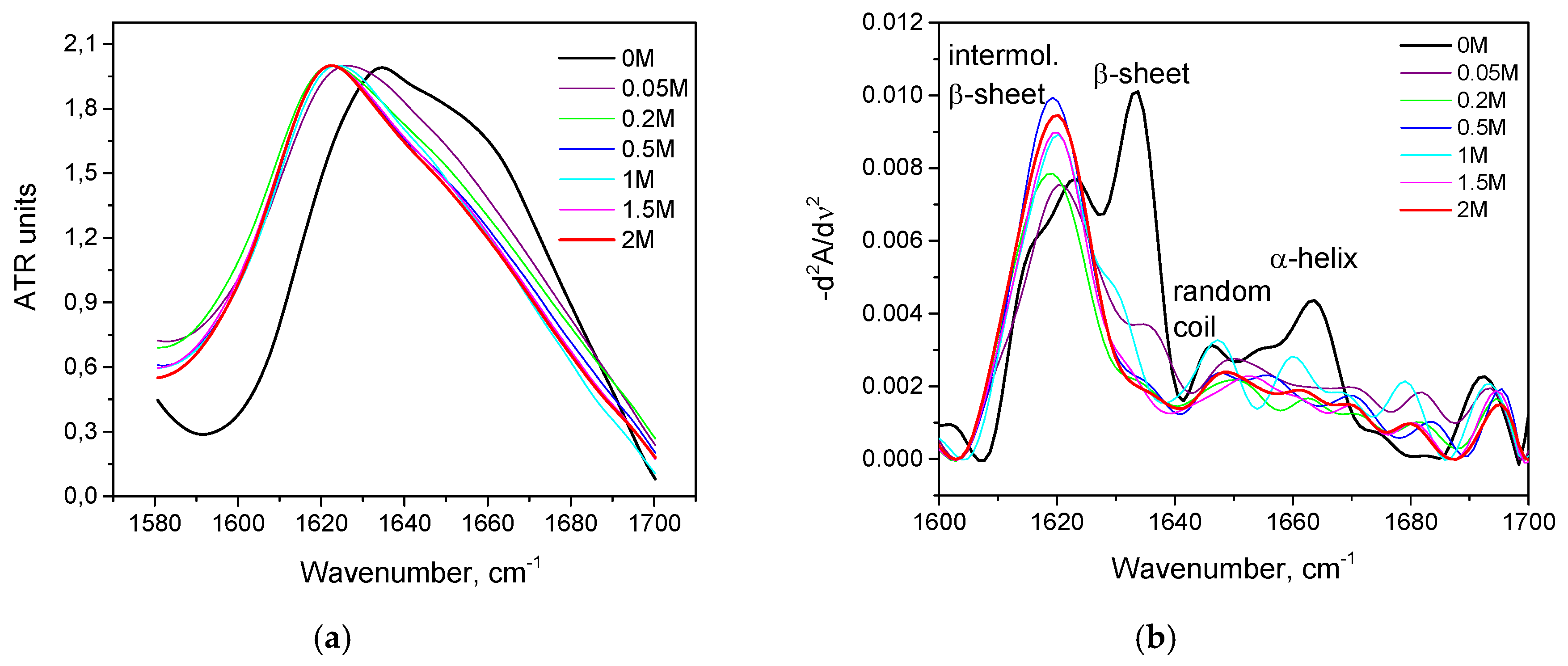
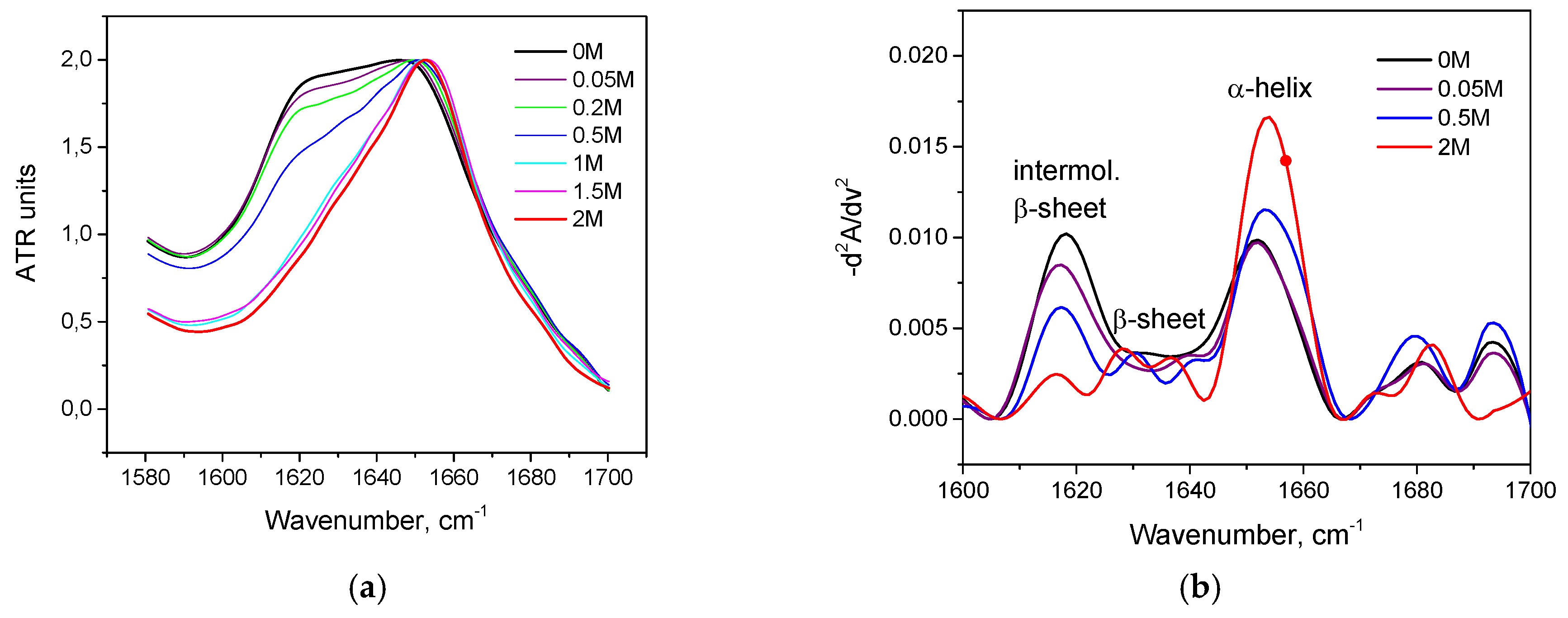
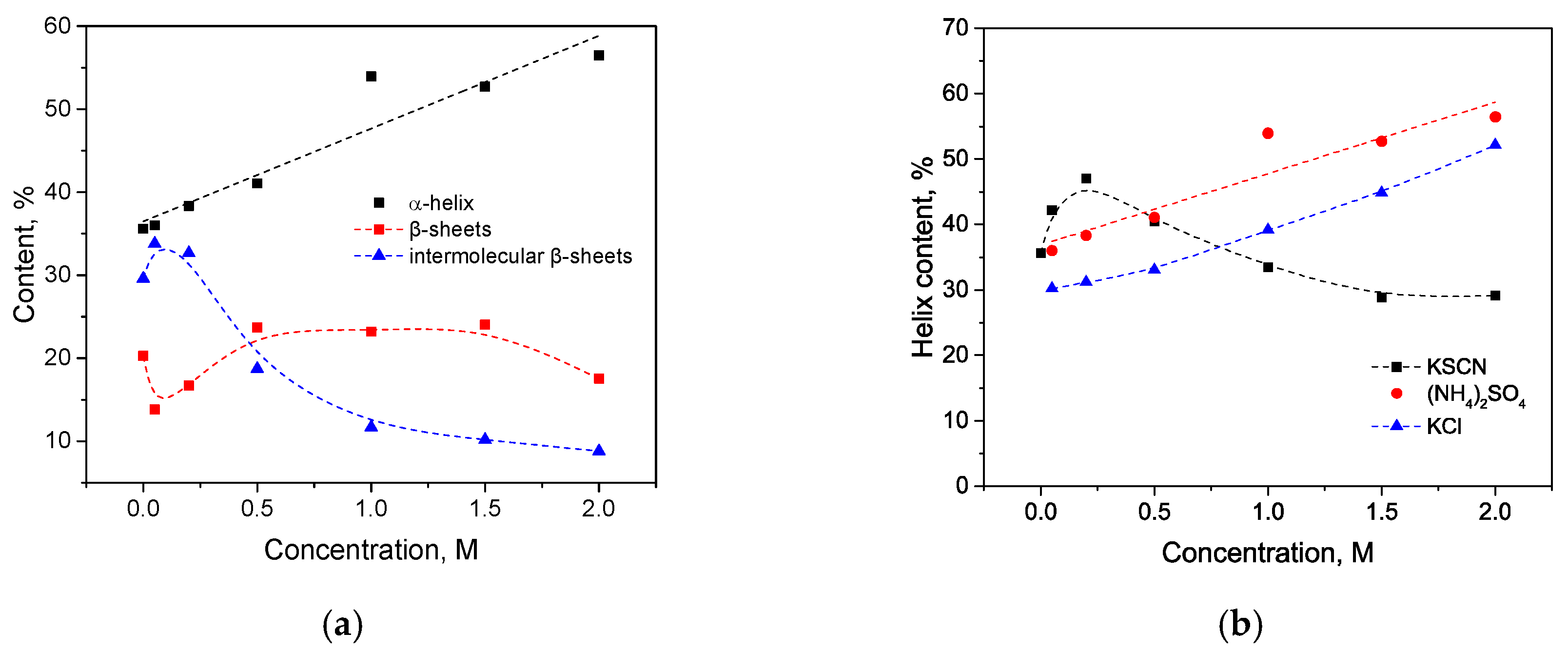
3.2. Salt-Induced Transition
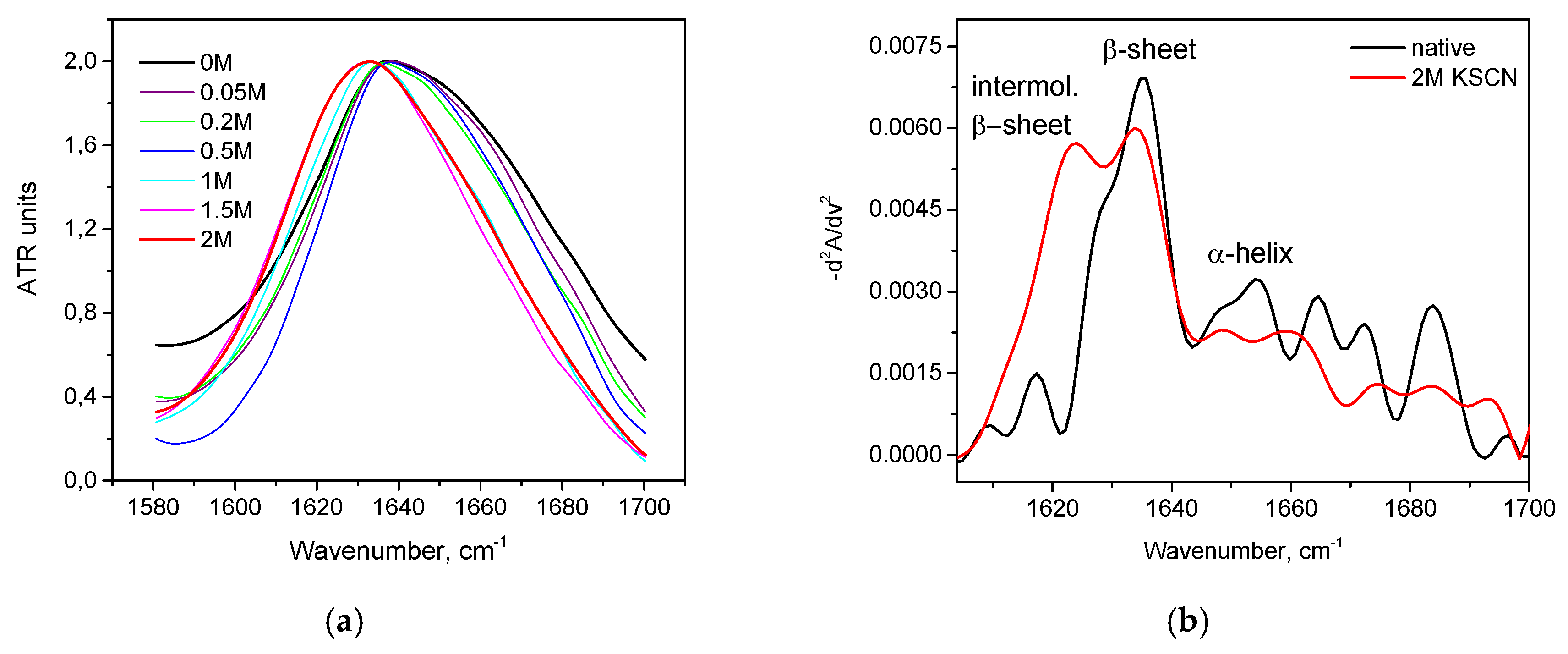
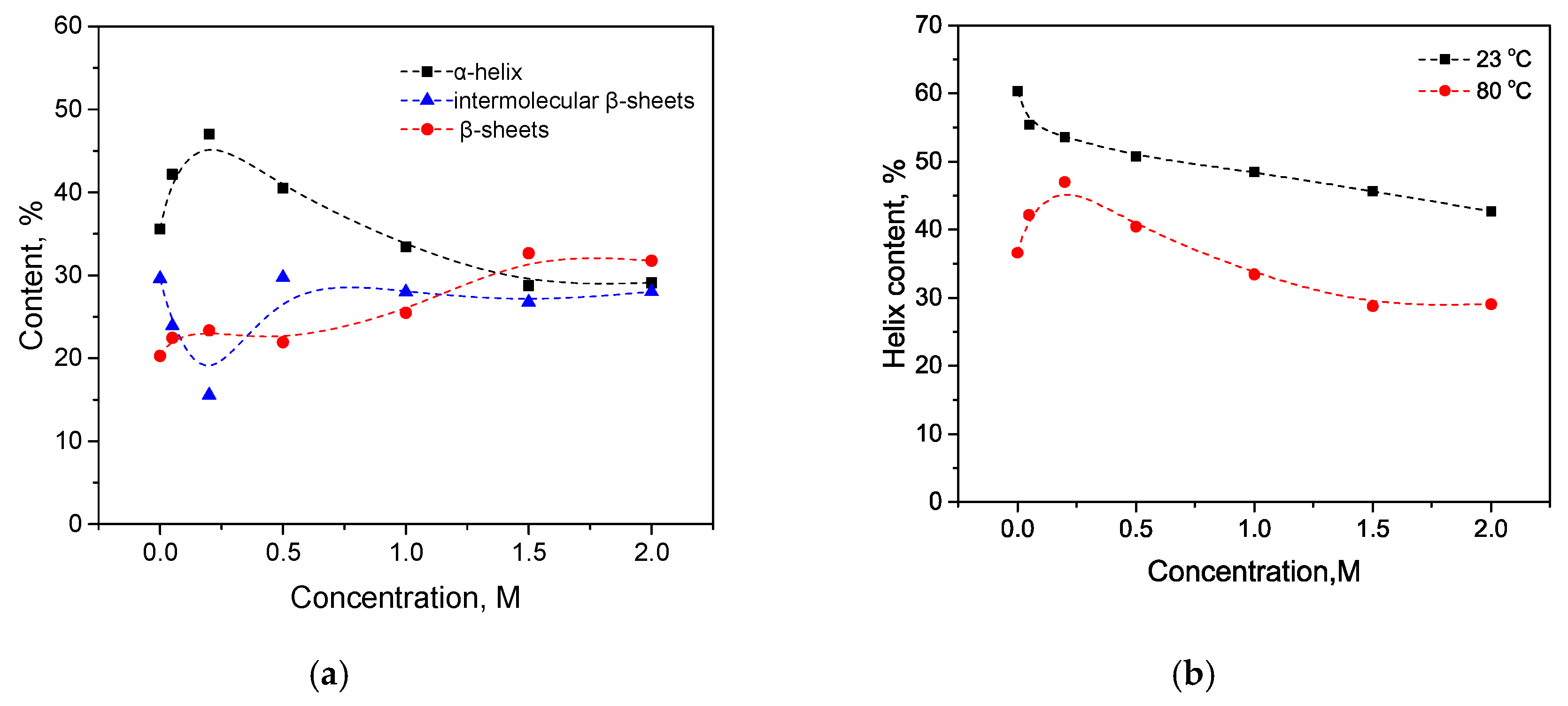
3.3. Combination of Salt and Temperature-Induced Transition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Lo Nostro, P.; Ninham, B.W. Hofmeister Phenomena: An Update on Ion Specificity in Biology. Chem. Rev. 2012, 112, 2286–2322. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H. Protein Stabilization and Enzyme Activation in Ionic Liquids: Specific Ion Effects. J. Chem. Technol. Biotechnol. 2016, 91, 25–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duong-Ly, K.C.; Gabelli, S.B. Salting out of Proteins Using Ammonium Sulfate Precipitation. Methods Enzymol. 2014, 541, 85–94. [Google Scholar] [PubMed]
- Zhang, R.; Li, N.; Liu, Y.; Han, X.; Tu, T.; Shen, J.; Xu, S.; Wu, Q.; Zhou, J.; Huang, Z. Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 β-xylosidase: Activity and instability at high neutral salt concentrations. Food Chem. 2019, 301, 125266. [Google Scholar] [CrossRef]
- Majumdar, R.; Manikwar, P.; Hickey, J.M.; Samra, H.S.; Sathish, H.A.; Bishop, S.M.; Middaugh, C.R.; Volkin, D.B.; Weis, D.D. Effects of Salts from the Hofmeister Series on the Conformational Stability, Aggregation Propensity, and Local Flexibility of an IgG1 Monoclonal Antibody. Biochemistry 2013, 52, 3376–3389. [Google Scholar] [CrossRef]
- Okur, H.I.; Hladílková, J.; Rembert, K.B.; Cho, Y.; Heyda, J.; Dzubiella, J.; Cremer, P.; Jungwirth, P. Beyond the Hofmeister Series: Ion-Specific Effects on Proteins and Their Biological Functions. J. Phys. Chem. B 2017, 121, 1997–2014. [Google Scholar] [CrossRef]
- Collins, K.D. Why Continuum Electrostatics Theories Cannot Explain Biological Structure, Polyelectrolytes or Ionic Strength Effects in Ion-protein Interactions. Biophys. Chem. 2012, 167, 43–59. [Google Scholar] [CrossRef] [Green Version]
- López-Arenas, L.; Solís-Mendiola, S.; Padilla-Zúñiga, J.; Hernández-Arana, A. Hofmeister effects in protein unfolding kinetics: Estimation of changes in surface area upon formation of the transition state. Biochim. Biophys. Acta 2006, 1764, 1260–1267. [Google Scholar] [CrossRef]
- Gokarn, Y.R.; Fesinmeyer, R.M.; Saluja, A.; Razinkov, V.; Chase, S.F.; Laue, T.M. Effective charge measurements reveal selective and preferential accumulation of anions, but not cations, at the protein surface in dilute salt solutions. Protein Sci. 2011, 20, 580–587. [Google Scholar] [CrossRef] [Green Version]
- Manciu, M.; Ruckenstein, E. Specific ion effects via ion hydration: I. Surface tension. Adv. Colloid Interface Sci. 2003, 105, 63–101. [Google Scholar] [CrossRef]
- Ioannou, F.; Leontidis, E.; Archontis, G. Helix Formation by Alanine-Based Peptides in Pure Water and Electrolyte Solutions: Insights from Molecular Dynamics Simulations. J. Phys. Chem. B 2013, 117, 9866–9876. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.B.; Strottmann, J.M.; Stellwagen, E. Prediction of neutral salt elution profiles for affinity-chromatography. Proc. Natl. Acad. Sci. USA 1981, 78, 2287–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dér, A.; Kelemen, L.; Fábián, L.; Taneva, S.G.; Fodor, E.; Páli, T.; Cupane, A.; Cacace, M.G.; Ramsden, J.I. Interfacial water structure controls protein conformation. J. Phys. Chem. B 2007, 111, 5344–5350. [Google Scholar] [CrossRef] [PubMed]
- Venkatesu, P.; Rani, A.; Kumar, A. A comparative study of the Hofmeister series of anions of the ionic salts and ionic liquids on the stability of α-chymotrypsin. New J. Chem. 2014, 39, 938–952. [Google Scholar]
- Andrea Salis, A.; Ninham, B.W. Models and mechanisms of Hofmeister effects in electrolyte solutions, and colloid and protein systems revisited. J. Chem. Soc. Rev. 2014, 43, 7358–7377. [Google Scholar] [CrossRef] [Green Version]
- Mazzini, V.; Craig, V.S.J. Volcano Plots Emerge from a Sea of Nonaqueous Solvents: The Law of Matching Water Affinities Extends to All Solvents. ACS Cent. Sci. 2018, 4, 1056–1064. [Google Scholar] [CrossRef]
- Collins, K.D. Ions from the Hofmeister series and osmolytes: Effects on proteins in solution and in the crystallization process. Methods 2004, 34, 300–311. [Google Scholar] [CrossRef]
- Collins, K.; Neilson, G.; Enderby, J. Ions in water: Characterizing the forces that control chemical processes and biological structure. Biophys. Chem. 2007, 128, 95–104. [Google Scholar] [CrossRef]
- Pace, C.N.; Prabhakaran, E.N.; Treviño, S.; Scholtz, J.M. Protein structure, stability and solubility in water and other solvents. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1225–1234. [Google Scholar] [CrossRef] [Green Version]
- Rembert, K.B.; Paterova, J.; Heyda, J.; Hilty, C.; Jungwirth, P.; Cremer, P.S. Molecular mechanisms of ion-specific effects on proteins. J. Am. Chem. Soc. 2012, 134, 10039–10046. [Google Scholar] [CrossRef]
- Medda, L.; Barse, B.; Cugia, F.; Boström, M.; Parsons, D.F.; Ninham, B.W.; Monduzzi, M.; Salis, A. Hofmeister Challenges: Ion Binding and Charge of the BSA Protein as Explicit Examples. Langmuir 2012, 28, 16355–16363. [Google Scholar] [CrossRef] [PubMed]
- Weingartner, H.; Cabrele, C.; Herrmann, C. How ionic liquids can help to stabilize native proteins. Phys. Chem. Chem. Phys. 2012, 14, 415–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cray, J.A.; Timson, D.J.; Russell, J.T.; Singhal, R.S. A universal measure of chaotropicity and kosmotropicity. Environ. Microbiol. 2013, 15, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Paterova, J.; Rembert, K.; Heyda, J.; Kurra, J.; Okur, H.; Liu, W.R.; Hilty, C.; Cremer, P.; Jungwirth, P. Reversal of the Hofmeister Series: Specific Ion Effects on Peptides. J. Phys. Chem. B 2013, 117, 8150–8158. [Google Scholar] [CrossRef] [PubMed]
- Arunkumar, R.; Drummond, C.J.; Greaves, T.L. FTIR Spectroscopic Study of the Secondary Structure of Globular Proteins in aqueous Protic Ionic Liquids. Front. Chem. 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Bostrom, M.; Parsons, D.F.; Salis, A.; Ninham, B.W.; Monduzzi, M. Possible Origin of the Inverse and Direct Hofmeister Series for Lysozyme at Low and High Salt Concentrations. Langmuir 2011, 27, 9504–9511. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L.; Frieden, C.; Rose, G.D. Dry Molten Globule Intermediates and the Mechanism of Protein Unfolding. Proteins 2010, 78, 2725–2737. [Google Scholar] [CrossRef] [Green Version]
- Crevenna, A.; Naredi-Rainer, N.; Lamb, D.C.; Wedlich-Söldner, R.; Dzubiella, J. Effects of Hofmeister Ions on the α-Helical Structure of Proteins. Biophys. J. 2012, 102, 907–915. [Google Scholar] [CrossRef] [Green Version]
- Hamada, D.; Kidokoro, S.; Fukada, H.; Takahashi, K.; Goto, Y. Salt-induced formation of the molten globule state of cytochrome c studied by isothermal titration calorimetry. Proc. Natl. Acad. Sci. USA 1994, 91, 10325–10329. [Google Scholar] [CrossRef] [Green Version]
- Goto, Y.; Adachi, M.; Muta, H.; So, M. Salt-induced formations of partially folded intermediates and amyloid fibrils suggests a common underlying mechanism. Biophys. Rev. 2018, 10, 493–502. [Google Scholar] [CrossRef] [Green Version]
- Usoltsev, D.; Sitnikova, V.; Kajava, A.; Uspenskaya, M. Systematic FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin under Various Denaturation Conditions. Biomolecules 2019, 9, 359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Susi, H.; Byler, D.M. Resolution-enhanced Fourier transform infrared spectroscopy of enzymes. Methods Enzymol. 1986, 130, 290–311. [Google Scholar] [CrossRef] [PubMed]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999, 12, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Deepthi, S.; Johnson, A.; Pattabhi, V. Structures of porcine beta-trypsin-detergent complexes: The stabilization of proteins through hydrophilic binding of polydocanol. Acta Crystallogr. Sect. D Boil. Crystallogr. 2000, 57, 1506–1512. [Google Scholar] [CrossRef] [Green Version]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen bonded and geometrical features. J. Biopolym. 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Touw, W.G.; Baakman, C.; Black, J.; te Beek, T.A.H.; Krieger, E.; Joosten, R.P.; Vriend, G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2014, 43, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Frishman, D.; Argos, P. Knowledge-Based Protein Secondary Structure Assignment Proteins: Structure, Function, and Genetics. Proteins 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Heinig, M.; Frishman, D. STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004, 32, W500–W502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulrichs, T.; Drotleff, A.M.; Ternes, W. Determination of heat-induced changes in the protein secondary structure of reconstituted livetins (water-soluble proteins from hen’s egg yolk) by FTIR. Food Chem. 2015, 172, 909–920. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Zhang, F.; Yu, S. Solid-film sampling method for the determination of protein secondary structure by Fourier transform infrared spectroscopy. Anal. Bioanal. Chem. 2017, 409, 4459–4465. [Google Scholar] [CrossRef] [PubMed]
- Yue Feng, Moyang Lv, YuQin Lu, Ke Liu, Lizhong Liu, Zhendan He, Kaimin Wu, Xinrong Wang, Baoshuang Zhang, Xuli Wu Characterization of binding interactions between selected phenylpropanoid glycosides and trypsin. Food Chem. 2018, 243, 118–124.
- Ahmed, A.B.; Znassi, N.; Chateau, M.; Kajava, A.V. A structure-based approach to predict predisposition to amyloidosis. Alzheimer’s Dement. 2015, 11, 681–690. [Google Scholar] [CrossRef]
- Yamasaki, M.; Yano, H.; Aoki, K. Differential scanning calorimetric studies on bovine serum albumin: II. Effects of neutral salts and urea. Int. J. Biol. Macromol. 1992, 13, 322–328. [Google Scholar] [CrossRef]
- Stirpe, A.; Pantusa, M.; Rizzuti, B.; De Santo, M.P.; Sportelli, L.; Bartucci, R.; Guzzi, R. Resveratrol induces thermal stabilization of human serum albumin and modulates the early aggregation stage. Int. J. Biol. Macromol. 2016, 92, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Shivu, B.; Seshadri, S.; Li, J.; Oberg, K.A.; Uversky, V.N.; Fink, A.L. Distinct β Sheet Structure in Protein Aggregates Determined by ATR−FTIR Spectroscopy. Biochemistry 2013, 52, 5176–5183. [Google Scholar] [CrossRef]
- Kumar Das, N.; Ghosh, N.; Kale, A.P.; Mondal, R.; Anand, U.; Ghosh, S.; Kumar Tiwari, V.; Kapur, M.; Mukherjee, S. Temperature Induced Morphological Transitions from Native to Unfolded Aggregated States of Human Serum Albumin. J. Phys. Chem. B 2014, 118, 7267–7276. [Google Scholar] [CrossRef]
- Capomaccio, R.; Osório, I.; Ojea-Jiménez, I.; Ceccone, G.; Colpo, P.; Gilliland, D.; Hussain, R.; Siligardi, G.; Rossi, F.; Ricard-Blum, S.; et al. Gold nanoparticles increases UV and thermal stability of human serum albumin. Biointerphases 2016, 11, 04B310. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Schubert, D.; Sawaya, M.R.; Eisenberg, D.; Riek, R. Multidimensional structure-activity relationship of a protein in its aggregated states. Angew. Chem. Int. Ed. 2010, 49, 3904–3908. [Google Scholar] [CrossRef]
- Kendrick, B.S.; Cleland, J.L.; Lam, X.; Nguyen, T.; Randolph, T.W.; Manning, M.C.; Carpenter, J.F. Aggregation of recombinant human interferon gamma: Kinetics and structural transition. J. Pharm. Sci. 1998, 87, 1069–1076. [Google Scholar] [CrossRef]









| Type | Ions | Jones–Dole Viscosity B Coefficient | Specific Effect on Water | Specific Effect on Protein |
|---|---|---|---|---|
| Cations | K+ | −0.007 | Chaotrope | stabilization |
| NH4+ | −0.007 | Chaotrope | stabilization | |
| Anions | SCN− | −0.103 | Chaotrope | denaturation |
| Cl− | −0.007 | Chaotrope | denaturation | |
| SO42− | 0.208 | Kosmotrope | stabilization |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usoltsev, D.; Sitnikova, V.; Kajava, A.; Uspenskaya, M. FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin and Trypsin under Neutral Salts. Biomolecules 2020, 10, 606. https://doi.org/10.3390/biom10040606
Usoltsev D, Sitnikova V, Kajava A, Uspenskaya M. FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin and Trypsin under Neutral Salts. Biomolecules. 2020; 10(4):606. https://doi.org/10.3390/biom10040606
Chicago/Turabian StyleUsoltsev, Dmitrii, Vera Sitnikova, Andrey Kajava, and Mayya Uspenskaya. 2020. "FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin and Trypsin under Neutral Salts" Biomolecules 10, no. 4: 606. https://doi.org/10.3390/biom10040606





