Activin A-Mediated Regulation of XT-I in Human Skin Fibroblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cytokine and Inhibitor Treatment
2.3. Nucleic Acid Extraction and Synthesis of Complementary DNA
2.4. Real-Time Polymerase Chain Reaction Analysis
2.5. Small Interfering RNA Transfection
2.6. Radiochemical Xylosyltransferase Activity Assay
2.7. Statistical Analysis
3. Results
3.1. Identification of Pro-Fibrotic Mediators Regulating XT-I Expression in Fibroblasts
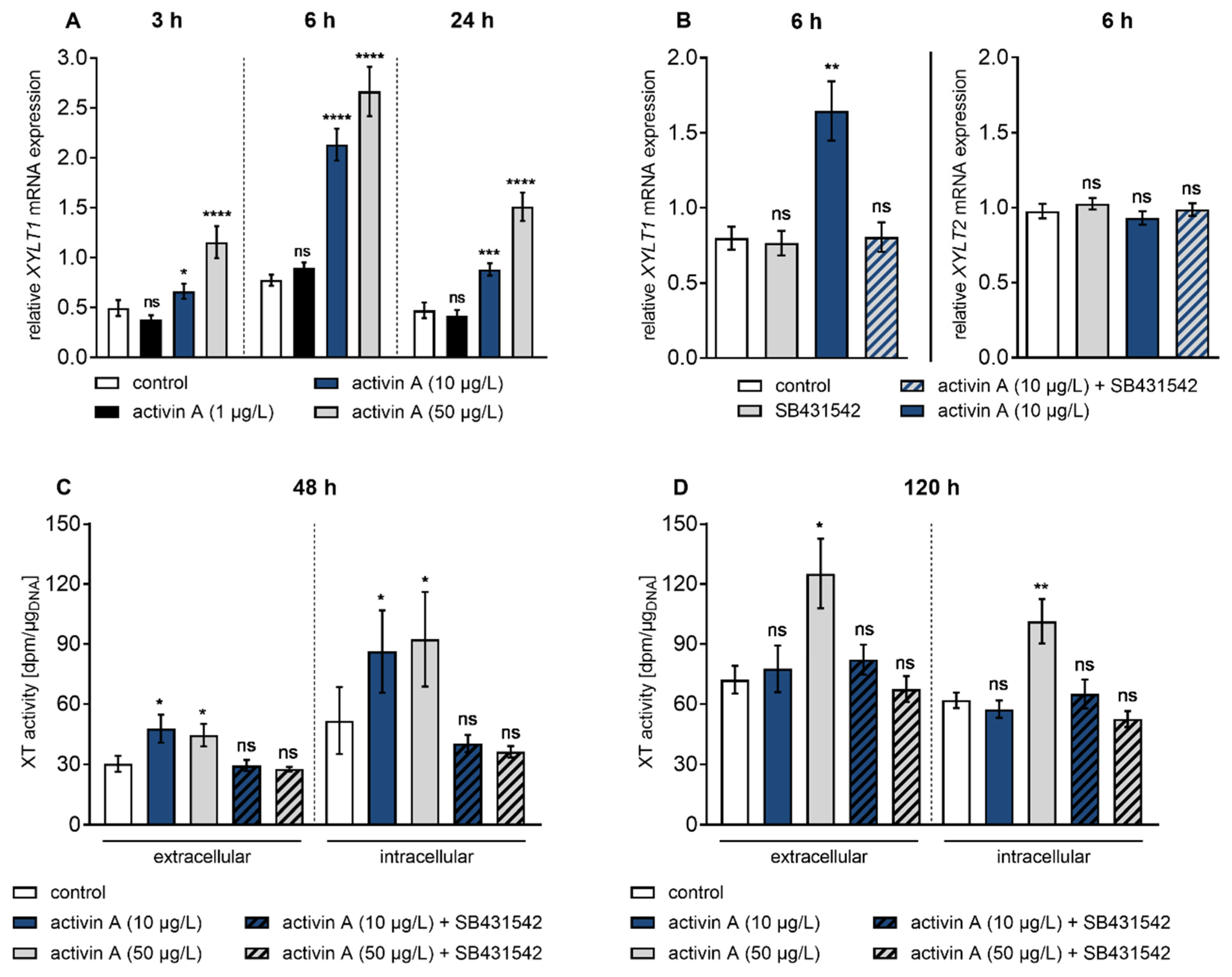
3.2. Activin A Induces Transient XYLT1 mRNA Expression and XT Activity Increase via ACVRIB/ALK4
3.3. ALK4–Activin A-Mediated XYLT1 mRNA Expression Requires MAPK Signalling Pathways
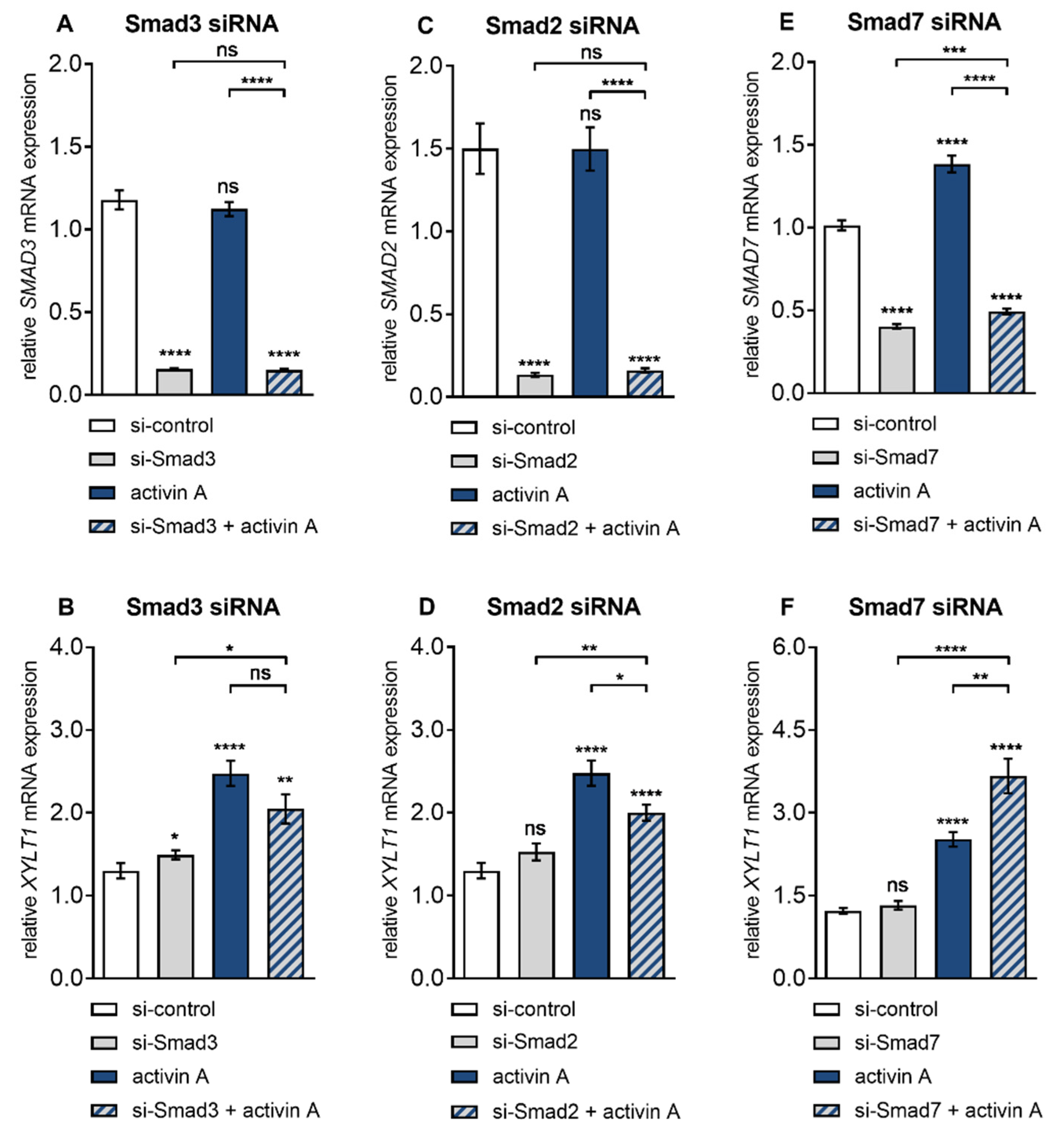
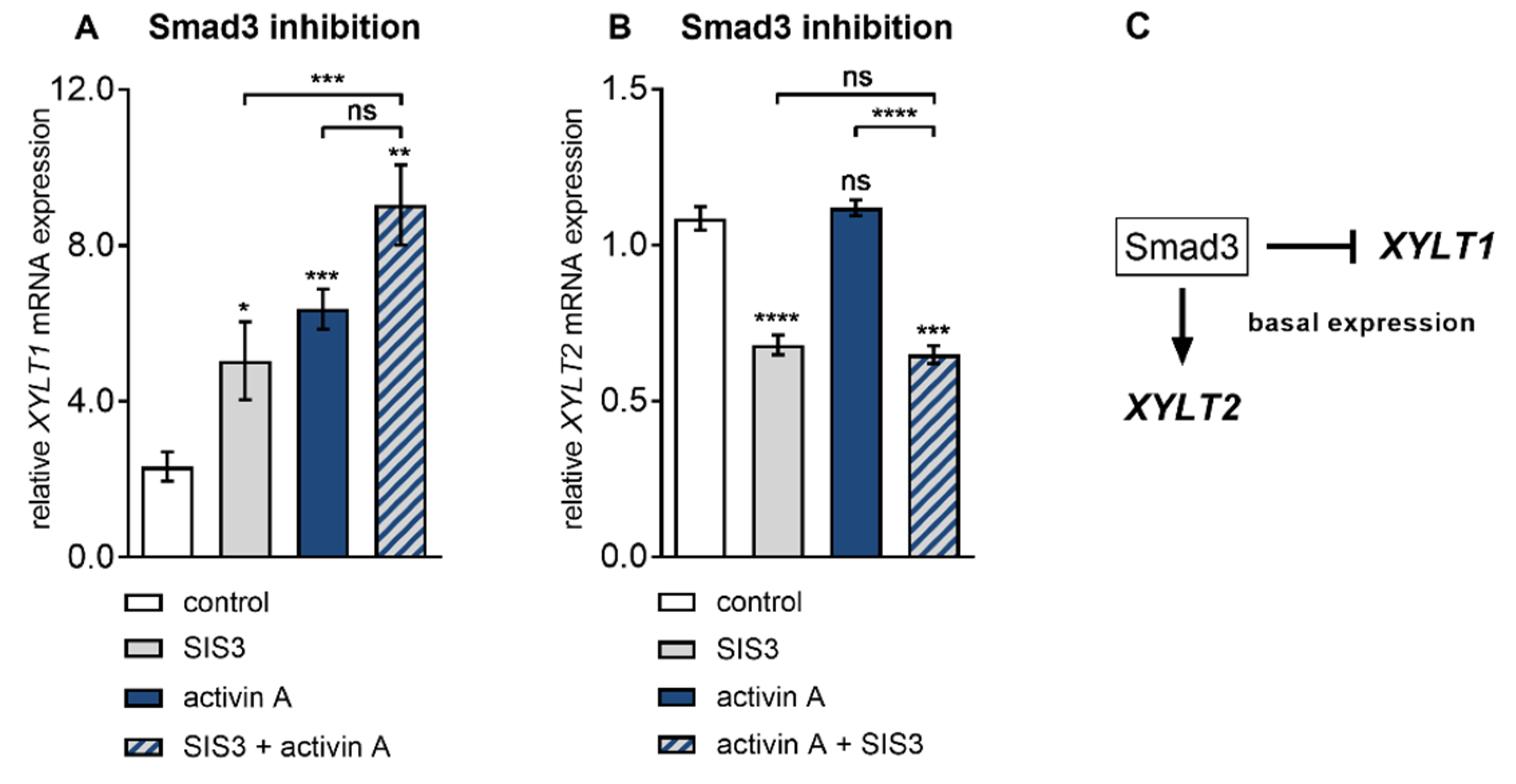
3.4. Smad3 is Dispensable for Activin A-Induced XYLT1 mRNA Expression in NHDF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014, 14, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Psarianos, P.; Ghoraie, L.S.; Yip, K.; Goldstein, D.; Gilbert, R.; Witterick, I.; Pang, H.; Hussain, A.; Lee, J.H.; et al. Metabolic regulation of dermal fibroblasts contributes to skin extracellular matrix homeostasis and fibrosis. Nat. Metab. 2019, 1, 147–157. [Google Scholar] [CrossRef]
- Do, N.N.; Eming, S.A. Skin fibrosis: Models and mechanisms. Curr. Res. Transl. Med. 2016, 64, 185–193. [Google Scholar] [CrossRef]
- Yazdani, S.; Bansal, R.; Prakash, J. Drug targeting to myofibroblasts: Implications for fibrosis and cancer. Adv. Drug Deliv. Rev. 2017, 121, 101–116. [Google Scholar] [CrossRef]
- Werner, S.; Smola, H. Paracrine regulation of keratinocyte proliferation and differentiation. Trends in Cell Biology 2001, 11, 143–146. [Google Scholar] [CrossRef]
- Borthwick, L.A.; Wynn, T.A.; Fisher, A.J. Cytokine mediated tissue fibrosis. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 1049–1060. [Google Scholar] [CrossRef] [Green Version]
- Ihn, H. Autocrine TGF-β signaling in the pathogenesis of systemic sclerosis. J. Dermatol. Sci. 2008, 49, 103–113. [Google Scholar] [CrossRef]
- Wang, X.; Fischer, G.; Hyvönen, M. Structure and activation of pro-activin A. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Annes, J.P. Making sense of latent TGFbeta activation. J. Cell Sci. 2003, 116, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Takagi, K.; Kawaguchi, Y.; Kawamoto, M.; Ota, Y.; Tochimoto, A.; Gono, T.; Katsumata, Y.; Takagi, M.; Hara, M.; Yamanaka, H. Activation of the Activin A-ALK-Smad pathway in systemic sclerosis. J. Autoimmun. 2011, 36, 181–188. [Google Scholar] [CrossRef]
- Jones, K.L.; De Kretser, D.M.; Patella, S.; Phillips, D.J. Activin A and follistatin in systemic inflammation. Mol. Cell. Endocrinol. 2004, 225, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Antsiferova, M.; Werner, S. The bright and the dark sides of activin in wound healing and cancer. J. Cell Sci. 2012, 125, 3929–3937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchida, K.; Nakatani, M.; Hitachi, K.; Uezumi, A.; Sunada, Y.; Ageta, H.; Inokuchi, K. Activin signaling as an emerging target for therapeutic interventions. Cell Commun. Signal. 2009, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fibroblasts, L.; Ohga, E.; Matsuse, T.; Teramoto, S.; Katayama, H.; Nagase, T.; Fukuchi, Y.; Ouchi, Y. Effects of Activin A on Proliferation and Differentiation Activin A is a dimeric protein that was isolated from ovarian fluid as a stimulator of follicle- stimulating hormone (FSH). Biochem. Biophys. Res. Commun. 1996, 228, 391–396. [Google Scholar]
- Yamashita, S.; Maeshima, A.; Kojima, I.; Nojima, Y. Activin A Is a Potent Activator of Renal Interstitial Fibroblasts. J. Am. Soc. Nephrol. 2004, 15, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Wang, X.; Wei, S.M.; Tang, Y.H.; Zhou, Q.; Huang, C.X. Activin A stimulates the proliferation and differentiation of cardiac fibroblasts via the ERK1/2 and p38-MAPK pathways. Eur. J. Pharmacol. 2016, 789, 319–327. [Google Scholar] [CrossRef]
- Bao, Y.L.; Tsuchida, K.; Liu, B.; Kurisaki, A.; Matsuzaki, T.; Sugino, H. Synergistic activity of activin A and basic fibroblast growth factor on tyrosine hydroxylase expression through Smad3 and ERK1/ERK2 MAPK signaling pathways. J. Endocrinol. 2005, 184, 493–504. [Google Scholar] [CrossRef] [Green Version]
- Susarla, B.T.S.S.; Laing, E.D.; Yu, P.; Katagiri, Y.; Geller, H.M.; Symes, A.J. Smad proteins differentially regulate transforming growth factor-β-mediated induction of chondroitin sulfate proteoglycans. J. Neurochem. 2011, 119, 868–878. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, S.; Chen, S.J.; Wu, M.; Warner-Blankenship, M.; Ning, H.; Lakos, G.; Mori, Y.; Chang, E.; Nihijima, C.; Takehara, K.; et al. Smad-independent transforming growth factor-β regulation of early growth response-1 and sustained expression in fibrosis: Implications for scleroderma. Am. J. Pathol. 2008, 173, 1085–1099. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Shimono, C.; Norioka, N.; Nakano, I.; Okubo, T.; Yagi, Y.; Hayashi, M.; Sato, Y.; Fujisaki, H.; Hattori, S.; et al. Activin A binds to perlecan through its pro-region that has heparin/heparan sulfate binding activity. J. Biol. Chem. 2010, 285, 36645–36655. [Google Scholar] [CrossRef] [Green Version]
- Kleesiek, K.; Reinards, R.; Okusi, J.; Wolf, B.; Greiling, H. UDP-D-Xylose: Proteoglycan Core Protein β-D-Xylosyltransferase: A New Marker of Cartilage Destruction in Chronic Joint Diseases. Clin. Chem. Lab. Med. 1987, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pönighaus, C.; Speirs, H.J.L.; Morris, B.J.; Kuhn, J.; Kleesiek, K.; Götting, C. Xylosyltransferase gene variants and their role in essential hypertension. Am. J. Hypertens. 2009, 22, 432–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götting, C.; Kuhn, J.; Zahn, R.; Brinkmann, T.; Kleesiek, K. Molecular Cloning and Expression of Human UDP-d-Xylose:Proteoglycan Core Protein β-d-Xylosyltransferase and its First Isoform XT-II. J. Mol. Biol. 2000, 304, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Pönighaus, C.; Kuhn, J.; Kleesiek, K.; Götting, C. Involvement of a cysteine protease in the secretion process of human xylosyltransferase I. Glycoconj. J. 2010, 27, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Götting, C.; Sollberg, S.; Kuhn, J.; Weilke, C.; Huerkamp, C.; Brinkmann, T.; Krieg, T.; Kleesick, K.; Kleesiek, K.; Sollberg, S.; et al. Serum xylosyltransferase: A new biochemical marker of the sclerotic process in systemic sclerosis. J. Investig. Dermatol. 1999, 112, 919–924. [Google Scholar] [CrossRef] [Green Version]
- Faust, I.; Roch, C.; Kuhn, J.; Prante, C.; Knabbe, C.; Hendig, D. Human xylosyltransferase-I – A new marker for myofibroblast differentiation in skin fibrosis. Biochem. Biophys. Res. Commun. 2013, 436, 449–454. [Google Scholar] [CrossRef]
- Prante, C.; Milting, H.; Kassner, A.; Farr, M.; Ambrosius, M.; Schon, S.; Seidler, D.G.; Banayosy, A.E.; Korfer, R.; Kuhn, J.; et al. Transforming Growth Factor beta1-regulated Xylosyltransferase I Activity in Human Cardiac Fibroblasts and Its Impact for Myocardial Remodeling. J. Biol. Chem. 2007, 282, 26441–26449. [Google Scholar] [CrossRef] [Green Version]
- Müller, B.; Prante, C.; Kleesiek, K.; Götting, C.; Muller, B.; Prante, C.; Kleesiek, K.; Gotting, C. Identification and Characterization of the Human Xylosyltransferase I Gene Promoter Region. J. Biol. Chem. 2009, 284, 30775–30782. [Google Scholar] [CrossRef] [Green Version]
- Faust, I.; Böker, K.O.; Lichtenberg, C.; Kuhn, J.; Knabbe, C.; Hendig, D. First description of the complete human xylosyltransferase-I promoter region. BMC Genet. 2014, 15, 1. [Google Scholar] [CrossRef] [Green Version]
- Müller, B.; Prante, C.; Knabbe, C.; Kleesiek, K.; Götting, C. First identification and functional analysis of the human xylosyltransferase II promoter. Glycoconj. J. 2013, 30, 237–245. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Götting, C.; Kuhn, J.; Tinneberg, H.-R.; Brinkmann, T.; Kleesiek, K. High xylosyltransferase activities in human follicular fluid and cultured granulosa-lutein cells. Mol. Hum. Reprod. 2002, 8, 1079–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Ishida, W.; Bhattacharyya, S.; Li, Y.; Platanias, L.C.; Varga, J. Selective inhibition of activin receptor-like kinase 5 signaling blocks profibrotic transforming growth factor β responses in skin fibroblasts. Arthr. Rheum. 2004, 50, 4008–4021. [Google Scholar] [CrossRef] [PubMed]
- Inman, G.J.; Nicolás, F.J.; Callahan, J.F.; Harling, J.D.; Gaster, L.M.; Reith, A.D.; Laping, N.J.; Hill, C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002, 62, 65–74. [Google Scholar] [CrossRef]
- Bennett, B.L.; Sasaki, D.T.; Murray, B.W.; Leary, E.C.O.; Sakata, S.T.; Xu, W.; Leisten, J.C.; Motiwala, A.; Pierce, S.; Satoh, Y.; et al. SP600125, an Anthrapyrazolone Inhibitor of Jun N-Terminal Kinase. Proc. Natl. Acad. Sci. USA 2001, 98, 13681–13686. [Google Scholar] [CrossRef] [Green Version]
- Henklova, P.; Vrzal, R.; Papouskova, B.; Bednar, P.; Jancova, P.; Anzenbacherova, E.; Ulrichova, J.; Maurel, P.; Pavek, P.; Dvorak, Z. SB203580, a pharmacological inhibitor of p38 MAP kinase transduction pathway activates ERK and JNK MAP kinases in primary cultures of human hepatocytes. Eur. J. Pharmacol. 2008, 593, 16–23. [Google Scholar] [CrossRef]
- Dumas, J. Protein kinase inhibitors: Emerging pharmacophores 1997 - 2000. Expert Opin. Ther. Pat. 2001, 11, 405–429. [Google Scholar] [CrossRef]
- Jinnin, M.; Ihn, H.; Tamaki, K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-β1-induced extracellular matrix expression. Mol. Pharmacol. 2006, 69, 597–607. [Google Scholar] [CrossRef]
- Kuhn, J.; Gressner, O.A.; Götting, C.; Gressner, A.M.; Kleesiek, K. Increased serum xylosyltransferase activity in patients with liver fibrosis. Clin. Chim. Acta 2009, 409, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Kitabatake, M.; Ishikawa, H.; Maeda, H. Immunohistochemical demonstration of proteoglycans in the skin of patients with systemic sclerosis. Br. J. Dermatol. 1983, 108, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Shinkai, H. Decorin and glycosaminoglycan synthesis in skin fibroblasts from patients with systemic sclerosis. Arch. Dermatol. Res. 1997, 289, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.L.; Johnson, K.E.; Harrison, C.A. Targeting TGF-β Mediated SMAD Signaling for the Prevention of Fibrosis. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khair, M.; Bourhim, M.; Barré, L.; Li, D.; Netter, P.; Magdalou, J.; Fournel-Gigleux, S.; Ouzzine, M. Regulation of Xylosyltransferase I Gene Expression by Interleukin 1β in Human Primary Chondrocyte Cells: MECHANISM AND IMPACT ON PROTEOGLYCAN SYNTHESIS. J. Biol. Chem. 2013, 288, 1774–1784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, W.; Zhou, J.; Markova, D.Z.; Tian, Y.; Li, J.; Anderson, D.G.; Shapiro, I.M.; Risbud, M.V. Xylosyltransferase-1 expression is refractory to inhibition by the inflammatory cytokines tumor necrosis factor α and IL-1β in nucleus pulposus cells: Novel regulation by AP-1, Sp1, and Sp3. Am. J. Pathol. 2014, 185, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Faust, I.; Donhauser, E.; Fischer, B.; Ibold, B.; Kuhn, J.; Knabbe, C.; Hendig, D. Characterization of dermal myofibroblast differentiation in pseudoxanthoma elasticum. Exp. Cell Res. 2017, 360, 153–162. [Google Scholar] [CrossRef]
- Fischer, B.; Ly, T.; Hendig, D.; Kuhn, J.; Pécheur, E.; Reungoat, E.; Knabbe, C.; Faust, I. Biochem. Biophys. Res. Commun. First description of a compensatory xylosyltransferase I induction observed after an anti fi brotic UDP-treatment of normal human dermal fi broblasts. Biochem. Biophys. Res. Commun. 2019, 512, 7–13. [Google Scholar] [CrossRef]
- Riedel, L.; Fischer, B.; Ly, T.-D.; Hendig, D.; Kuhn, J.; Knabbe, C.; Faust, I. microRNA-29b mediates fibrotic induction of human xylosyltransferase-I in human dermal fibroblasts via the Sp1 pathway. Sci. Rep. 2018, 8, 17779. [Google Scholar] [CrossRef] [Green Version]
- Garrett, S.M.; Baker Frost, D.; Feghali-Bostwick, C. The Mighty Fibroblast and Its Utility in Scleroderma Research. J. Scleroderma Relat. Disord. 2017, 2, 100–107. [Google Scholar] [CrossRef]
- Kamato, D.; Burch, M.; Zhou, Y.; Mohamed, R.; Stow, J.L.; Osman, N.; Zheng, W.; Little, P.J. Individual Smad2 linker region phosphorylation sites determine the expression of proteoglycan and glycosaminoglycan synthesizing genes. Cell. Signal. 2019, 53, 365–373. [Google Scholar] [CrossRef] [Green Version]
- Meurer, S.K.; Weiskirchen, R. Usage of Mitogen-activated protein kinase small molecule inhibitors: More than just inhibition! Front. Pharmacol. 2018, 9, 9–11. [Google Scholar] [CrossRef] [Green Version]
- Dolivo, D.M.; Larson, S.A.; Dominko, T. Crosstalk between mitogen-activated protein kinase inhibitors and transforming growth factor-β signaling results in variable activation of human dermal fibroblasts. Int. J. Mol. Med. 2019, 43, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Kempf, H.; Lecina, M.; Ting, S.; Zweigerdt, R.; Oh, S. Distinct regulation of mitogen-activated protein kinase activities is coupled with enhanced cardiac differentiation of human embryonic stem cells. Stem Cell Res. 2011, 7, 198–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotokezaka, H.; Sakai, E.; Kanaoka, K.; Saito, K.; Matsuo, K.; Kitaura, H.; Yoshida, N.; Nakayama, K. U0126 and PD98059, Specific Inhibitors of MEK, Accelerate Differentiation of RAW264.7 Cells into Osteoclast-like Cells. J. Biol. Chem. 2002, 277, 47366–47372. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Kobayashi, T.; Funatsu, O.; Morita, A.; Ikekita, M. Biochem. Biophys. Res. Commun. Activin A induces neuronal differentiation and survival via ALK4 in a SMAD-independent manner in a subpopulation of human neuroblastomas. Biochem. Biophys. Res. Commun. 2010, 394, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Rostam, M.A.; Shajimoon, A.; Kamato, D.; Mitra, P.; Piva, T.J.; Getachew, R.; Cao, Y.; Zheng, W.; Osman, N.; Little, P.J. Flavopiridol Inhibits TGF- β -Stimulated Biglycan Synthesis by Blocking Linker Region Phosphorylation and Nuclear Translocation of Smad2. J. Pharmacol. Exp. Ther. 2018, 365, 156–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rostam, M.A.; Kamato, D.; Piva, T.J.; Zheng, W.; Little, P.J.; Osman, N. The role of specific Smad linker region phosphorylation in TGF-β mediated expression of glycosaminoglycan synthesizing enzymes in vascular smooth muscle. Cell. Signal. 2016, 28, 956–966. [Google Scholar] [CrossRef]
- Mori, Y.; Chen, S.J.; Varga, J. Expression and regulation of intracellular SMAD signaling in scleroderma skin fibroblasts. Arthr. Rheum. 2003, 48, 1964–1978. [Google Scholar] [CrossRef]
- Venkatesan, N.; Barré, L.; Bourhim, M.; Magdalou, J.; Mainard, D.; Netter, P.; Fournel-Gigleux, S.; Ouzzine, M. Xylosyltransferase-I Regulates Glycosaminoglycan Synthesis during the Pathogenic Process of Human Osteoarthritis. PLoS ONE 2012, 7, e34020. [Google Scholar] [CrossRef]
- Ihn, H.; Yamane, K.; Asano, Y.; Jinnin, M.; Tamaki, K. Constitutively phosphorylated Smad3 interacts with Sp1 and p300 in scleroderma fibroblasts. Rheumatology 2006, 45, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, S.; Sargent, J.L.; Du, P.; Lin, S.; Tourtellotte, W.G.; Takehara, K.; Whitfield, M.L.; Varga, J. Egr-1 induces a profibrotic injury/repair gene program associated with systemic sclerosis. PLoS ONE 2011, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Piera-velazquez, S.; Makul, A.; Jimenez, S.A. Increased Expression of NAPDH Oxidase 4 (NOX4) in Systemic Sclerosis Dermal Fibroblasts: Regulation by Transforming Growth Factor β. Arthr. Rheumatol. 2016, 67, 2749–2758. [Google Scholar]
- Tan, N.Y.; Khachigian, L.M. Sp1 Phosphorylation and Its Regulation of Gene Transcription. Mol. Cell. Biol. 2009, 29, 2483–2488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, K.R.; Corey, D.A.; Dunn, J.M.; Kelley, T.J. SMAD3 expression is regulated by mitogen-activated protein kinase kinase-1 in epithelial and smooth muscle cells. Cellular Signalling 2007, 19, 923–931. [Google Scholar] [CrossRef]
- Poncelet, A.-C.; De Caestecker, M.P.; Schnaper, H.W. The transforming growth factor-βbgr/SMAD signaling pathway is present and functional in human mesangial cells. Kidney Int. 1999, 56, 1354–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baugé, C.; Cauvard, O.; Leclercq, S.; Galéra, P.; Boumédiene, K. Modulation of transforming growth factor beta signalling pathway genes by transforming growth factor beta in human osteoarthritic chondrocytes: Involvement of Sp1 in both early and late response cells to transforming growth factor beta. Arthr. Res. Ther. 2011, 13, R23. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Massagué, J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [Green Version]
- Ungefroren, H.; Groth, S.; Sebens, S.; Lehnert, H.; Gieseler, F.; Fändrich, F. Differential roles of Smad2 and Smad3 in the regulation of TGF-β1-mediated growth inhibition and cell migration in pancreatic ductal adenocarcinoma cells: Control by Rac1. Mol. Cancer 2011, 10, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | Primers | TA (°C) | Product Size (bp) |
|---|---|---|---|
| ACTA2 | 5′-GACCGAATGCAGAAGGAG-3′ 5′-CGGTGGACAATGGAAGG-3′ | 59 | 169 |
| B2M | 5‘-TGTGCTCGCGCTACTCTCTCTT-3‘ 5′-CGGATGGATGAAACCCAGACA-3′ | 59 | 137 |
| RPL13A | 5′-CGGAAGGTGGTGGTCGTA-3′ 5′-CTCGGGAAGGGTTGGTGT-3′ | 63 | 165 |
| SDHA | 5′-AACTCGCTCTTGGACCTG-3′ 5′-GAGTCGCAGTTCCGATGT-3′ | 63 | 177 |
| SMAD2 | 5′-ACAACAGGCCTTTACAGCTTCT-3′ 5′-GGAGGCAAAACTGGTGTCTCA-3′ | 63 | 239 |
| SMAD3 | 5′-ACCATCCGCATGAGCTTC-3′ 5′-CACTGCAAAGGCCCATTC-3′ | 63 | 107 |
| SMAD7 | 5′-AGATGCTGTGCCTTCCTC-3′ 5′-GTCTTCTCCTCCCAGTATGC-3′ | 63 | 135 |
| TGFB1 | 5′-GCGATACCTCAGCAACC-3′ 5′-ACGCAGCAGTTCTTCTCC-3′ | 59 | 331 |
| XYLT1 | 5′-GAAGCCGTGGTGAATCAG-3′ 5′-CGGTCAGCAAGGAAGTAG-3′ | 63 | 281 |
| XYLT2 | 5′-ACACAGATGACCCGCTTGTGG-3′ 5′-TTGGTGACCCGCAGGTTGTTG-3′ | 63 | 139 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ly, T.-D.; Plümers, R.; Fischer, B.; Schmidt, V.; Hendig, D.; Kuhn, J.; Knabbe, C.; Faust, I. Activin A-Mediated Regulation of XT-I in Human Skin Fibroblasts. Biomolecules 2020, 10, 609. https://doi.org/10.3390/biom10040609
Ly T-D, Plümers R, Fischer B, Schmidt V, Hendig D, Kuhn J, Knabbe C, Faust I. Activin A-Mediated Regulation of XT-I in Human Skin Fibroblasts. Biomolecules. 2020; 10(4):609. https://doi.org/10.3390/biom10040609
Chicago/Turabian StyleLy, Thanh-Diep, Ricarda Plümers, Bastian Fischer, Vanessa Schmidt, Doris Hendig, Joachim Kuhn, Cornelius Knabbe, and Isabel Faust. 2020. "Activin A-Mediated Regulation of XT-I in Human Skin Fibroblasts" Biomolecules 10, no. 4: 609. https://doi.org/10.3390/biom10040609






