The MAP Kinase Phosphatase MKP-1 Modulates Neurogenesis via Effects on BNIP3 and Autophagy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Immunohistochemistry (IHC)
2.3. Neural Stem Cells
2.4. Quantitative PCR
2.5. Immunocytochemistry
2.6. Immunoblotting
2.7. Flow Cytometry
2.8. Luciferase Assays
2.9. siRNA Transfection
2.10. Statistical Analysis
3. Results
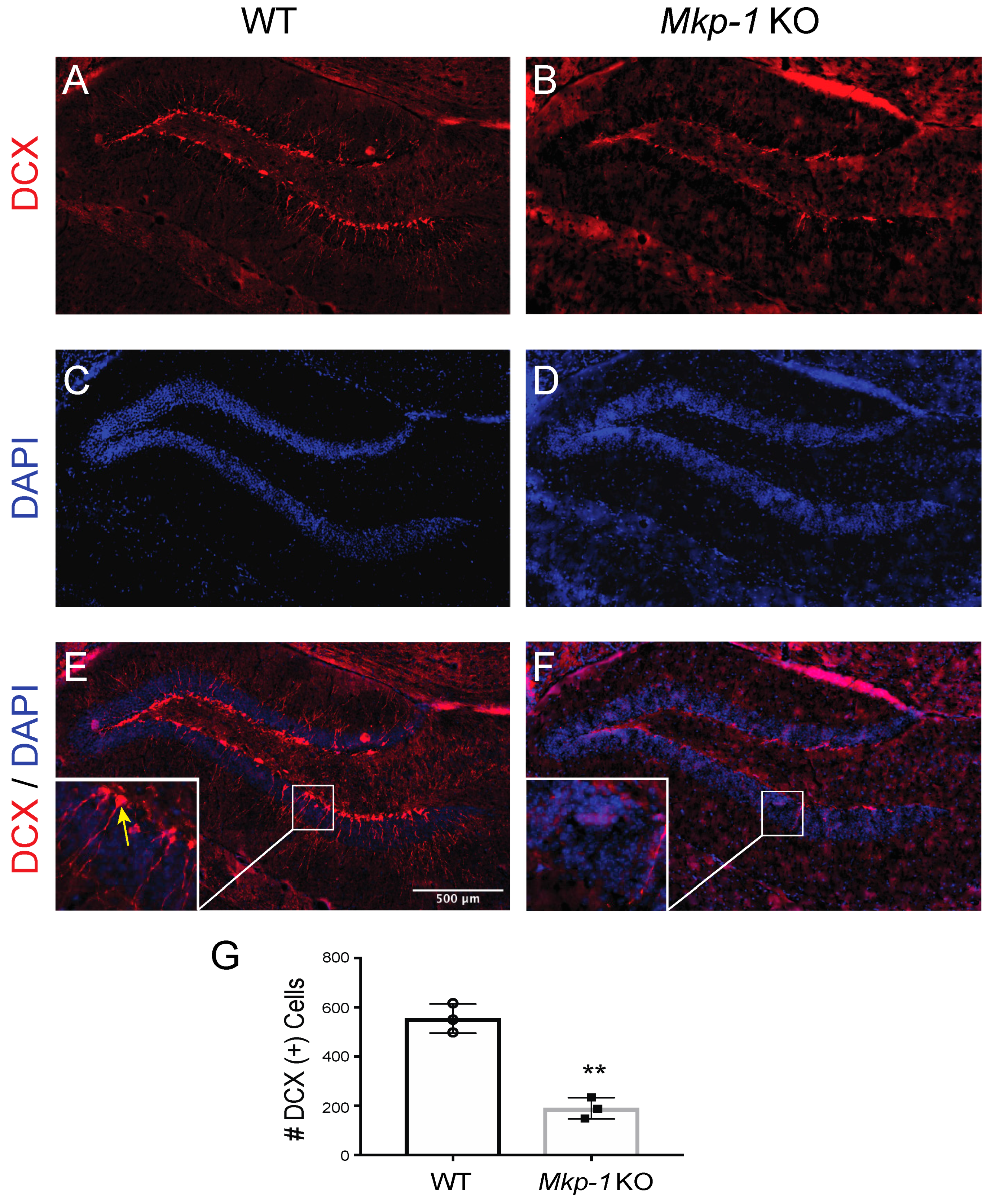
3.1. Loss of MKP-1 Has a Suppressive Effect on Neurogenesis
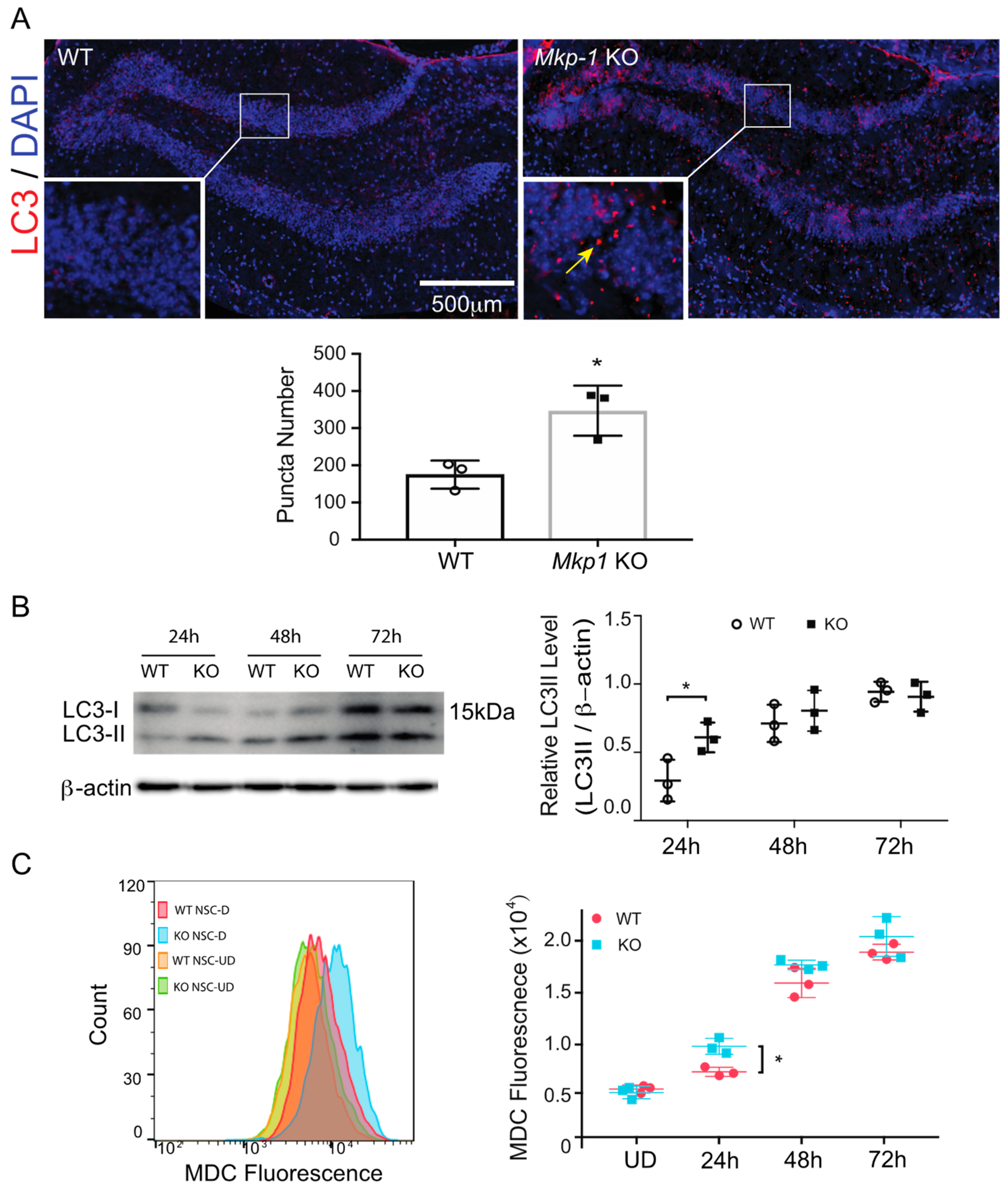
3.2. Loss of MKP-1 Induces Autophagy in Differentiated NSCs
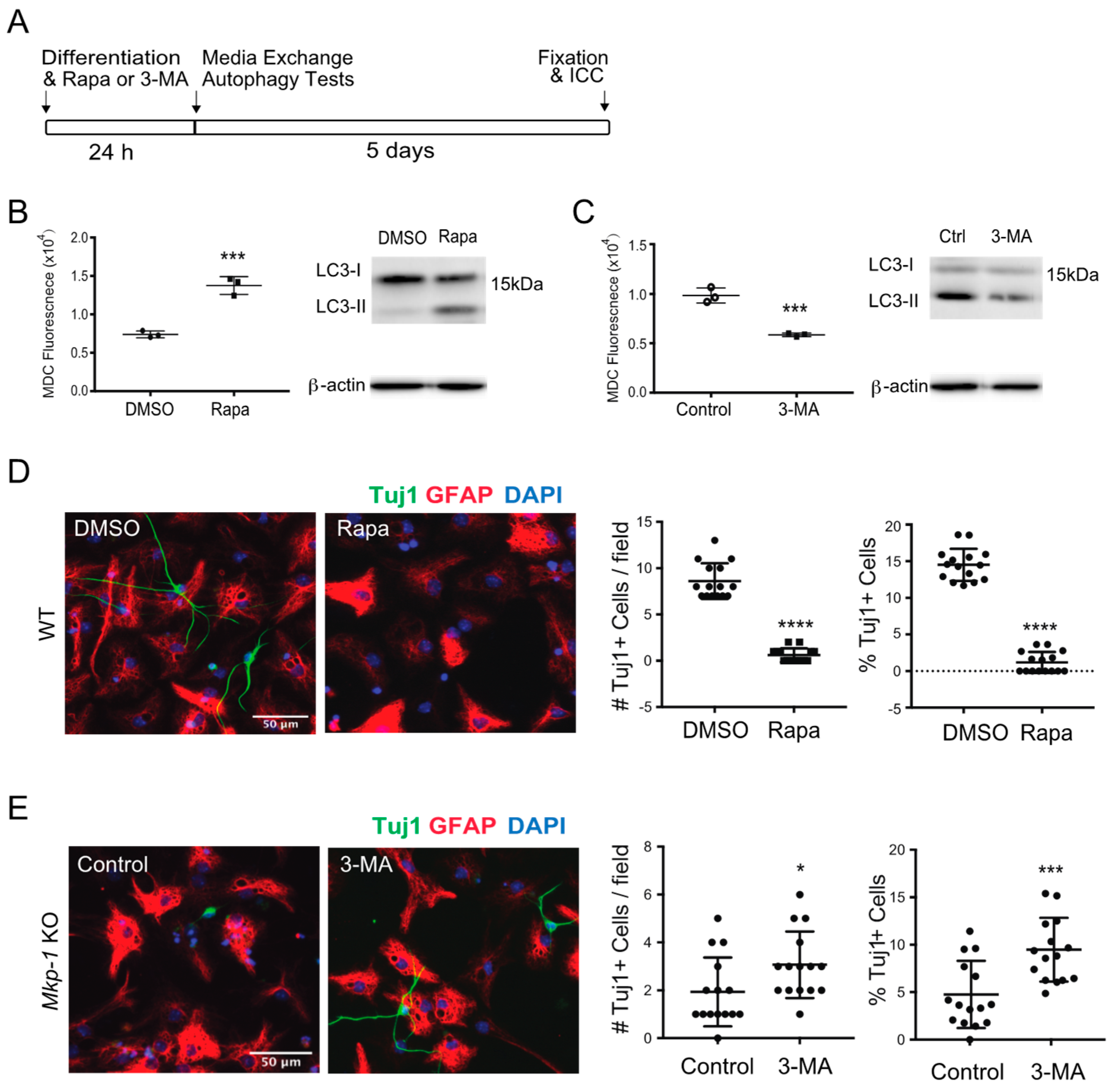
3.3. Effect of MKP-1 Deficiency on Neurogenesis Is Autophagy-Dependent
3.4. BNIP3 Regulates Autophagy and Neurogenesis in Mkp-1 KO NSCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blows, W.T. Child brain development. Nurs. Times 2003, 99, 28–31. [Google Scholar]
- Patzke, N.; Spocter, M.A.; Karlsson, K.; Bertelsen, M.F.; Haagensen, M.; Chawana, R.; Streicher, S.; Kaswera, C.; Gilissen, E.; Alagaili, A.N.; et al. In contrast to many other mammals, cetaceans have relatively small hippocampi that appear to lack adult neurogenesis. Brain Struct. Funct. 2013, 220, 361–383. [Google Scholar] [CrossRef]
- Palmer, T.D.; Willhoite, A.R.; Gage, F.H. Vascular niche for adult hippocampal neurogenesis. J. Comp. Neurol. 2000, 42, 479–494. [Google Scholar] [CrossRef]
- Spalding, K.; Bergmann, O.; Alkass, K.; Bernard, S.; Salehpour, M.; Huttner, H.B.; Boström, E.; Westerlund, I.; Vial, C.; Buchholz, B.; et al. Dynamics of Hippocampal Neurogenesis in Adult Humans. Cell 2013, 153, 1219–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.Z.; Nusslock, R. Exercise-Mediated Neurogenesis in the Hippocampus via BDNF. Front. Neurosci. 2018, 12, 52. [Google Scholar] [CrossRef] [Green Version]
- Artegiani, B.; Calegari, F. Age-related cognitive decline: Can neural stem cells help us? Aging 2012, 4, 176–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheyuo, C.; Aziz, M.; Wang, P. Neurogenesis in Neurodegenerative Diseases: Role of MFG-E8. Front. Neurosci. 2019, 13, 569. [Google Scholar] [CrossRef]
- Nakano-Kobayashi, A.; Awaya, T.; Kii, I.; Sumida, Y.; Okuno, Y.; Yoshida, S.; Sumida, T.; Inoue, H.; Hosoya, T.; Hagiwara, M. Prenatal neurogenesis induction therapy normalizes brain structure and function in Down syndrome mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10268–10273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngwenya, L.; Danzer, S.C. Impact of Traumatic Brain Injury on Neurogenesis. Front. Neurosci. 2019, 12, 1014. [Google Scholar] [CrossRef]
- Lu, J.; Manaenko, A.; Hu, Q. Targeting Adult Neurogenesis for Poststroke Therapy. Stem Cells Int. 2017, 2017, 5868632. [Google Scholar] [CrossRef] [Green Version]
- Ravikumar, B.; Sarkar, S.; Davies, J.E.; Futter, M.; Garcia-Arencibia, M.; Green-Thompson, Z.W.; Jimenez-Sanchez, M.; Korolchuk, V.; Lichtenberg, M.; Luo, S.; et al. Regulation of Mammalian Autophagy in Physiology and Pathophysiology. Physiol. Rev. 2010, 90, 1383–1435. [Google Scholar] [CrossRef] [Green Version]
- Wirawan, E.; Berghe, T.V.; Lippens, S.; Agostinis, P.; Vandenabeele, P. Autophagy: For better or for worse. Cell Res. 2011, 22, 43–61. [Google Scholar] [CrossRef]
- Parzych, K.R.; Klionsky, D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casares-Crespo, L.; Calatayud-Baselga, I.; García-Corzo, L.; Mira, H. On the Role of Basal Autophagy in Adult Neural Stem Cells and Neurogenesis. Front. Cell. Neurosci. 2018, 12, 339. [Google Scholar] [CrossRef]
- Zapata-Muñoz, J.; Villarejo-Zori, B.; Largo-Barrientos, P.; Boya, P. Towards a better understanding of the neuro-developmental role of autophagy in sickness and in health. Cell Stress 2021, 5, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Y. Regulation of innate immune response by MAP kinase phosphatase-1. Cell. Signal. 2007, 19, 1372–1382. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhou, J.-Y.; Kho, D.; Reiners, J.J.R., Jr.; Wu, G.S. Role for DUSP1 (dual-specificity protein phosphatase 1) in the regulation of autophagy. Autophagy 2016, 12, 1791–1803. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.-H.; Chen, C.-Z.; Li, S.; Han, D.-X.; Wang, Y.-J.; Yuan, B.; Gao, Y.; Zhang, J.-B.; Jiang, H. Dual-specificity phosphatase 1 regulates cell cycle progression and apoptosis in cumulus cells by affecting mitochondrial function, oxidative stress, and autophagy. Am. J. Physiol. Physiol. 2019, 317, C1183–C1193. [Google Scholar] [CrossRef]
- Jin, Q.; Li, R.; Hu, N.; Xin, T.; Zhu, P.; Hu, S.; Ma, S.; Zhu, H.; Ren, J.; Zhou, H. DUSP1 alleviates cardiac ischemia/reperfusion injury by suppressing the Mff-required mitochondrial fission and Bnip3-related mitophagy via the JNK pathways. Redox Biol. 2017, 14, 576–587. [Google Scholar] [CrossRef]
- Du, Y.; Du, Y.; Zhang, Y.; Huang, Z.; Fu, M.; Li, J.; Pang, Y.; Lei, P.; Wang, Y.T.; Song, W.; et al. MKP-1 reduces Aβ generation and alleviates cognitive impairments in Alzheimer’s disease models. Signal Transduct. Target. Ther. 2019, 4, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, D.M.; Moser, R.; Régulier, E.; Breuillaud, L.; Dixon, M.; Beesen, A.A.; Elliston, L.; Santos, M.D.F.S.; Kim, J.; Jones, L.; et al. MAP kinase phosphatase 1 (MKP-1/DUSP1) is neuroprotective in Huntington’s disease via additive effects of JNK and p38 inhibition. J. Neurosci. 2013, 33, 2313–2325. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Liu, Y.; Sun, M. Autophagy and Alzheimer’s Disease. Cell Mol. Neurobiol. 2017, 37, 377–388. [Google Scholar] [CrossRef]
- Croce, K.R.; Yamamoto, A. A role for autophagy in Huntington’s disease. Neurobiol. Dis. 2019, 122, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yi, P.; Zhang, B.; Xu, C.; Liu, Q.; Pi, Z.; Xu, X.; Chevet, E.; Liu, J. Differences in endoplasmic reticulum stress signalling kinetics determine cell survival outcome through activation of MKP-1. Cell. Signal. 2011, 23, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.J.; Wang, G.C.; Zuo, F.X.; Li, X.Y.; Wu, J.; Chen, G.; Dou, W.C.; Guo, Y.; Shen, Q.; Wang, R.Z. Transcriptome profiling of the subventricular zone and dentate gyrus in an animal model of Parkinson’s disease. Int. J. Mol. Med. 2017, 40, 771–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, B.N.; Albert, G.P.; Halterman, M.W. Expression Profiling of the MAP Kinase Phosphatase Family Reveals a Role for DUSP1 in the Glioblastoma Stem Cell Niche. Cancer Microenviron. 2017, 10, 57–68. [Google Scholar] [CrossRef]
- Wu, J.J.; Bennett, A.M. Essential Role for Mitogen-activated Protein (MAP) Kinase Phosphatase-1 in Stress-responsive MAP Kinase and Cell Survival Signaling. J. Biol. Chem. 2005, 280, 16461–16466. [Google Scholar] [CrossRef] [Green Version]
- Azari, H.; Sharififar, S.; Rahman, M.; Ansari, S.; Reynolds, B.A. Establishing Embryonic Mouse Neural Stem Cell Culture Using the Neurosphere Assay. J. Vis. Exp. 2011, 47, e2457. [Google Scholar] [CrossRef] [Green Version]
- Selvaraj, P.; Xiao, L.; Lee, C.; Murthy, S.R.; Cawley, N.X.; Lane, M.; Merchenthaler, I.; Ahn, S.; Loh, Y.P. Neurotrophic Factor-α1: A Key Wnt-β-Catenin Dependent Anti-Proliferation Factor and ERK-Sox9 Activated Inducer of Embryonic Neural Stem Cell Differentiation to Astrocytes in Neurodevelopment. Stem Cells 2017, 35, 557–571. [Google Scholar] [CrossRef]
- Biederbick, A.; Kern, H.F.; Elsässer, H.P. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 1995, 66, 3–14. [Google Scholar]
- Bruick, R.K. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA 2000, 97, 9082–9087. [Google Scholar] [CrossRef] [Green Version]
- Rahman, N.Z.A.; Greenwood, S.M.; Brett, R.R.; Tossell, K.; Ungless, M.A.; Plevin, R.; Bushell, T.J. Mitogen-Activated Protein Kinase Phosphatase-2 Deletion Impairs Synaptic Plasticity and Hippocampal-Dependent Memory. J. Neurosci. 2016, 36, 2348–2354. [Google Scholar] [CrossRef] [Green Version]
- Collins, L.M.; Downer, E.J.; Toulouse, A.; Nolan, Y.M. Mitogen-Activated Protein Kinase Phosphatase (MKP)-1 in Nervous System Development and Disease. Mol. Neurobiol. 2014, 51, 1158–1167. [Google Scholar] [CrossRef]
- Finelli, M.J.; Murphy, K.J.; Chen, L.; Zou, H. Differential Phosphorylation of Smad1 Integrates BMP and Neurotrophin Pathways through Erk/Dusp in Axon Development. Cell Rep. 2013, 3, 1592–1606. [Google Scholar] [CrossRef] [Green Version]
- Guan, J.-L.; Simon, A.K.; Prescott, M.; Menendez, J.A.; Liu, F.; Wang, F.; Wang, C.; Wolvetang, E.; Vazquez-Martin, A.; Zhang, J. Autophagy in stem cells. Autophagy 2013, 9, 830–849. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-R.; Hu, H.-Y.; Kuo, S.-H.; Lei, H.-Y.; Lin, Y.-S.; Yeh, T.-M.; Liu, C.-C.; Liu, H.-S. Dengue virus infection induces autophagy: An in vivo study. J. Biomed. Sci. 2013, 20, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, R.-Y.; Zhu, S.-H.; Li, V.; Gibson, S.; Xu, X.; Kong, J.-M. BNIP3 Interacting with LC3 Triggers Excessive Mitophagy in Delayed Neuronal Death in Stroke. CNS Neurosci. Ther. 2014, 20, 1045–1055. [Google Scholar] [CrossRef]
- Collins, L.M.; O’Keeffe, G.W.; Long-Smith, C.M.; Wyatt, S.L.; Sullivan, A.M.; Toulouse, A.; Nolan, Y.M. Mitogen-Activated Protein Kinase Phosphatase (MKP)-1 as a Neuroprotective Agent: Promotion of the Morphological Development of Midbrain Dopaminergic Neurons. Neuro Mol. Med. 2013, 15, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Koga, S.; Kojima, S.; Kishimoto, T.; Kuwabara, S.; Yamaguchi, A. Over-expression of map kinase phosphatase-1 (MKP-1) suppresses neuronal death through regulating JNK signaling in hypoxia/re-oxygenation. Brain Res. 2012, 1436, 137–146. [Google Scholar] [CrossRef]
- Ryan, S.M.; Nolan, Y.M. Neuroinflammation negatively affects adult hippocampal neurogenesis and cognition: Can exercise compensate? Neurosci. Biobehav. Rev. 2016, 61, 121–131. [Google Scholar] [CrossRef]
- Roux, C.; Lesueur, C.; Aligny, C.; Brasse-Lagnel, C.; Genty, D.; Marret, S.; Laquerrière, A.; Bekri, S.; Gonzalez, B.J. 3-MA Inhibits Autophagy and Favors Long-Term Integration of Grafted Gad67–GFP GABAergic Precursors in the Developing Neocortex by Preventing Apoptosis. Cell Transplant. 2014, 23, 1425–1450. [Google Scholar] [CrossRef] [Green Version]
- De Gioia, R.; Biella, F.; Citterio, G.; Rizzo, F.; Abati, E.; Nizzardo, M.; Bresolin, N.; Comi, G.P.; Corti, S. Neural Stem Cell Transplantation for Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 3103. [Google Scholar] [CrossRef] [PubMed]
- Chrostek, M.; Fellows, E.G.; Crane, A.T.; Grande, A.; Low, W.C. Efficacy of stem cell-based therapies for stroke. Brain Res. 2019, 1722, 146362. [Google Scholar] [CrossRef] [PubMed]
- Daido, S.; Kanzawa, T.; Yamamoto, A.; Takeuchi, H.; Kondo, Y.; Kondo, S. Pivotal Role of the Cell Death Factor BNIP3 in Ceramide-Induced Autophagic Cell Death in Malignant Glioma Cells. Cancer Res. 2004, 64, 4286–4293. [Google Scholar] [CrossRef] [Green Version]
- Ott, C.; König, J.; Höhn, A.; Jung, T.; Grune, T. Macroautophagy is impaired in old murine brain tissue as well as in senescent human fibroblasts. Redox Biol. 2016, 10, 266–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Shi, Y.; Han, Z.; Pan, Y.; Liu, N.; Han, S.; Chen, Y.; Lan, M.; Qiao, T.; Fan, D. Suppression of the dual-specificity phosphatase MKP-1 enhances HIF-1 trans-activation and increases expression of EPO. Biochem. Biophys. Res. Commun. 2003, 312, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Talwar, H.; Bauerfeld, C.; Bouhamdan, M.; Farshi, P.; Liu, Y.; Samavati, L. MKP-1 negatively regulates LPS-mediated IL-1β production through p38 activation and HIF-1α expression. Cell. Signal. 2017, 34, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y. Role of HIF-1a in regulating autophagic cell survival during cerebral ischemia reperfusion in rats. Oncotarget 2017, 8, 98482–98494. [Google Scholar] [CrossRef] [Green Version]
- Gao, A.; Jiang, J.; Xie, F.; Chen, L. Bnip3 in mitophagy: Novel insights and potential therapeutic target for diseases of secondary mitochondrial dysfunction. Clin. Chim. Acta 2020, 506, 72–83. [Google Scholar] [CrossRef]
- Ney, P.A. Mitochondrial autophagy: Origins, significance, and role of BNIP3 and NIX. Biochim. Biophys. Acta Bioenergy 2015, 1853, 2775–2783. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, Y.; Kim, E.; Beemiller, P.; Wang, C.-Y.; Swanson, J.; You, M.; Guan, K.-L. Bnip3 Mediates the Hypoxia-induced Inhibition on Mammalian Target of Rapamycin by Interacting with Rheb. J. Biol. Chem. 2007, 282, 35803–35813. [Google Scholar] [CrossRef] [Green Version]
- Rubinstein, A.D.; Kimchi, A. Life in the balance—A mechanistic view of the crosstalk between autophagy and apoptosis. J. Cell Sci. 2012, 125, 5259–5268. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.Z.; Westberg, J.A.; Holtta, E.; Andersson, L.C. BCL2 regulates neural differentiation. Proc. Natl. Acad. Sci. USA 1996, 93, 4504–4508. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, T.; Kitagawa, K.; Yagita, Y.; Sugiura, S.; Omura-Matsuoka, E.; Tanaka, S.; Matsushita, K.; Okano, H.; Tsujimoto, Y.; Hori, M. Bcl2 enhances survival of newborn neurons in the normal and ischemic hippocampus. J. Neurosci. Res. 2006, 84, 1187–1196. [Google Scholar] [CrossRef]
- Bernier, P.J.; Parent, A. Bcl-2 Protein as a Marker of Neuronal Immaturity in Postnatal Primate Brain. J. Neurosci. 1998, 18, 2486–2497. [Google Scholar] [CrossRef] [Green Version]
- Bernier, P.J.; Vinet, J.; Cossette, M.; Parent, A. Characterization of the subventricular zone of the adult human brain: Evidence for the involvement of Bcl-2. Neurosci. Res. 2000, 37, 67–78. [Google Scholar] [CrossRef]
- Chang, M.Y.; Sun, W.; Ochiai, W.; Nakashima, K.; Kim, S.Y.; Park, C.H.; Kang, J.S.; Shim, J.W.; Jo, A.Y.; Kang, C.S.; et al. Bcl-XL/Bax proteins direct the fate of embryonic cortical precursor cells. Mol. Cell. Biol. 2007, 27, 4293–4305. [Google Scholar] [CrossRef] [Green Version]
- Ray, R.; Chen, G.; Velde, C.V.; Cizeau, J.; Park, J.H.; Reed, J.C.; Gietz, R.D.; Greenberg, A.H. BNIP3 heterodimerizes with Bcl-2/Bcl-XL and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 2000, 275, 1439–1448. [Google Scholar] [CrossRef] [Green Version]
- Moosavi, S.M.; Prabhala, P.; Ammit, A.J. Role and regulation of MKP-1 in airway inflammation. Respir. Res. 2017, 18, 154. [Google Scholar] [CrossRef]
- Liu, L.; Doran, S.; Xu, Y.; Manwani, B.; Ritzel, R.; Benashski, S.; McCullough, L.; Li, J. Inhibition of mitogen-activated protein kinase phosphatase-1 (MKP-1) increases experimental stroke injury. Exp. Neurol. 2014, 261, 404–411. [Google Scholar] [CrossRef]
- Aelvoet, S.-A.; Pascual-Brazo, J.; Libbrecht, S.; Reumers, V.; Gijsbers, R.; Haute, C.V.D.; Baekelandt, V. Long-Term Fate Mapping Using Conditional Lentiviral Vectors Reveals a Continuous Contribution of Radial Glia-Like Cells to Adult Hippocampal Neurogenesis in Mice. PLoS ONE 2015, 10, e0143772. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Halterman, M.W. The MAP Kinase Phosphatase MKP-1 Modulates Neurogenesis via Effects on BNIP3 and Autophagy. Biomolecules 2021, 11, 1871. https://doi.org/10.3390/biom11121871
Li Y, Halterman MW. The MAP Kinase Phosphatase MKP-1 Modulates Neurogenesis via Effects on BNIP3 and Autophagy. Biomolecules. 2021; 11(12):1871. https://doi.org/10.3390/biom11121871
Chicago/Turabian StyleLi, Yinghui, and Marc W. Halterman. 2021. "The MAP Kinase Phosphatase MKP-1 Modulates Neurogenesis via Effects on BNIP3 and Autophagy" Biomolecules 11, no. 12: 1871. https://doi.org/10.3390/biom11121871





