Computational Approaches to the Rational Design of Tubulin-Targeting Agents
Abstract
:1. Introduction
2. Ligand-Based Approaches
2.1. Similarity Search
2.2. QSAR Modeling
2.3. Pharmacophore Screening
3. Structure-Based Approaches
3.1. Structural Data on Tubulin
3.1.1. Tools to Study Tubulin 3D Structures
3.1.2. Binding Sites on Tubulin
3.1.3. System Selection for Virtual Screening (VS) and MD Simulations
3.2. Tubulin-Related VS Strategies
3.2.1. Pharmacophore Screening
3.2.2. Protein-Ligand Docking
3.3. Molecular Dynamics (MD) Simulations to Study Tubulin-Ligand Complexes
3.3.1. Classical MD Simulations Used on Tubulin
3.3.2. Enhanced Sampling Methods
4. Umbrella Sampling (US)
5. Steered Molecular Dynamics Simulations (SMD)
6. Metadynamics (MetaD)
7. Applications of MD for Tubulin-Ligand Studies
7.1. Docking Validation and Refinement
7.2. Comparison of the Binding Free Energy of Different Ligands
7.3. Identification of Key Binding Site Residues
7.4. Analysis of Local and Global Effects upon Ligand Binding
7.5. Exploration of Ligand Binding to Different Tubulin Isotypes
7.6. MD Analysis Metrics
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Reference | Screened Dataset | Dataset Size | Software | Descriptors | Similarity Metric | Result |
|---|---|---|---|---|---|---|
| Ayoub 2013 [16] | PubChem | Built-in web search | 881-bit PubChem subgraph fingerprints [URL] | Tanimoto, 80% similarity threshold | Virtual hits ranked by protein-ligand docking, one compound used as a reference for further successful design | |
| Guo 2019 [17] | ChemDiv | Discovery Studio (Biovia) | ECFP-4 | Tanimoto, 50% similarity threshold | Virtual hits were found to be cytotoxic, one was confirmed as a colchicine site binder | |
| Lo 2015 [20] | ChEMBL, PubChem | CSNAP2D | OpenBabel FP2 | Tanimoto, 85% similarity threshold | Correctly identified and validated tubulin as a target for 36 molecules that showed cytotoxicity in a HTS setting | |
| Lo 2016 [21] | CSNAP3D | ShapeAlign | 3D Tanimoto, 85% similarity threshold | A virtual hit was established to promote tubulin polymerization by binding at the taxane site | ||
| Magiatordi [18] | CoCoCo | Phase (Schrödin-ger) | Atom-type-based 3D shape | Atom-type volume scoring, 0.65 similarity threshold | 31 virtual hits have been confirmed to decrease microtubule polymerization in vitro. | |
| Federico 2020 [19] | ZINC *, Chembridge Diverset CL, Chembridge Diverset EXP, BindingDB FDA, MayBridge | ROCS (OpenEye) | Smooth 3D Gaussian functions for each atom | Tanimoto similarities of aligned overlap volumes (no threshold, top 5000 selected for ROCS, top 2000 for EON) | Two virtual hits established by shape and electrostatic similarity to a known active were shown to inhibit tubulin polymerization in vitro. | |
| EON (OpenEye) | Electrostatic potential maps of pre-aligned molecules |
| Reference | Modeled Data | Descriptor Type | Algorithm | Validation Strategy | Application and Result |
|---|---|---|---|---|---|
| Gaikwad 2018 [25] | IC50 of 102 phenylindoles cytotoxic against MCF7 cancer cell line | Fragment-based holograms implemented in SYBYL-X (Certara) | PLS | Two sets were used: training (77) and test (25). Leave- one-out and five-fold cross-validation were used. | Analysis of literature data allowed the authors to highlight structural features important for cytotoxicity. |
| Extended connectivity fingerprints, physicochemical descriptors | Naïve Bayes (Discovery Studio 3.0, Accelrys) | ||||
| Guo 2020 [26] | 1076 diverse colchicine-site targeting small molecules extracted from the ChEMBL database | Extended-connectivity fingerprints, path-based fingerprints | Naïve Bayes | Five-fold cross-validation. | A colchicine site-binding inhibitor of tubulin polymerization was established after a virtual screening campaign. |
| Single Tree | |||||
| Random Forest | |||||
| Stefanski 2018 [27] | IC50 of 83 thio-derivatives of combretastatin-A4 mined from literature | Extended connectivity fingerprints, physicochemical descriptors | Naïve Bayes | Leave-one-out, cross-validation, and external test set methods. The external validation test set was composed of 20 tubulin inhibitors and 800 decoys. | Two virtual hits selected by consensus QSAR modeling were later confirmed to be cytotoxic due to perturbing microtubule polymerization by binding at the colchicine site. |
| Multiple Linear Regression | |||||
| Quan 2018 [28] | IC50 values of 64 literature-mined derivatives of combretastatin A-4 | CoMFA (steric and electrostatic fields) | PLS (SYBYL-X 2.0, Tripos) | Leave-one-out validation | A 3D QSAR study highlighted structural elements with pronounced relation to activity value, useful for further optimization. |
| CoMSIA (steric, electrostatic, hydrophobic, hydrogen bond donor, and hydrogen bond acceptor fields) | |||||
| Pandit 2021 [29] | IC50 values of 49 tubulysin derivatives reported in the literature | CoMFA (steric and electrostatic fields) | PLS (SYBYL-X 2.0, Tripos) | Cross-validation | 3D QSAR investigation of structure-activity data on tubulysins lead to rational design and synthesis of a new class of cytotoxic in vitro tubulysin derivatives |
| CoMSIA (steric, electrostatic, hydrophobic, hydrogen bond donor, and hydrogen bond acceptor fields) |
| Reference | Compound Library | Compound Set Used to Build the Model | Software Used to Build the Model | Model Generation and Validation Settings | Validation Set | Validation Metric and Score | Screening Result |
|---|---|---|---|---|---|---|---|
| Zhang 2021 [32] | BioDiversity, 30,000 molecules | Six agents targeting taxane site | HipHop algorithm from Discovery Studio 3.5 (Accelrys) | Five features were used (HBA, HBD, HP, HP-A, and R-A) 1, paclitaxel used as reference | 467 inactive molecules from ZINC15 database, 33 known inhibitors | Gunner-Henry (GH) score of 0.62 | Large database filtered to focus on a subset that eventually led to discovery of two taxane-site targeting cytotoxic agents |
| Lone 2017 [33] | IBScreen Natural Product Database, 84,215 molecules | Four C20 substituted vinblastine analogues extracted from literature | Phase (Schrödinger) | HBA, HBD, HP, PI, and R-A 1 | 35 inactive and four active C20 substituted vinblastine analogues | The Survival-inactive score of 4.006. | Possibility of scaffold-hopping for vinca-site-targeting compounds design was shown |
| Niu 2014 [34] | Specs Screening Database, 202,919 molecules | 26 compounds designed to target colchicine site with known cytotoxic action | HypoGen module from Discovery Studio 2.5 (Accelrys) | HBD, HBA, HP, and R-A 1 | 66 colchicine site-targeting compounds with known cytotoxicity (26 actives, 40 inactives) | Cost difference | Two compounds with good fitness to the developed pharmaco-phore model were shown to be tubulin polymerization inhibitors in vitro. |
| Stefanski 2018 [27] | A custom-designed virtual combinatorial library of 1159 combretastatin A-4 analogs | 21 active colchicine site-targeting molecules mined from literature | Discovery Studio 3.5 (Accelrys) | HBA, HBD, HP, and HP-A 1 | 20 tubulin inhibitors and 800 decoys mined from ChEMBL | Area under receiver-operator curve (AUROC) | Two virtual hits were established as in vitro cytotoxic agents targeting colchicine binding site |
| Reference | Data Used to Build the Model | Software | Validation Set | Screened Data | Result |
|---|---|---|---|---|---|
| Nagarajan 2015 [82] | 1SA0, 1SA1, 3HKC, 3HKE, 3HKD, 3N2K, 3N2G; model derived from 1SA0 was manually removed of a hydrogen bond feature, shown best result in validation | Model building: LigandScout v3.1 (Inte:Ligand); Screening—Phase v3.4 (Schrödinger) | 52 active colchicine site binders mined from literature, 1800 decoy molecules from the DUD database | The CoCoCo database, containing multiconformer data on 3.7 million purchasable compounds | 31 novel colchicine site-targeting inhibitors of tubulin polymerization were established that match the derived pharmacophore model |
| Mangiatordi 2017 [18] | 6F7C, 5EYP, 5YL2, 4O2B (common feature model) | MOE (Chemical Computing Group Inc.) | 970 inactive molecules and 30 known inhibitors with experimental activity mined from literature | Specs database, 202,919 molecules | The screening established five virtual hits that are cytotoxic in vitro, one most potent hit confirmed to bind at the colchicine site |
| Zhou 2019 [83] | 118 crystal structures of tubulin co-crystalized with colchicine site binding ligands | LigandScout v3.1 (Inte:Ligand), Phase v3.4 (Schrödinger), Pharmer | 81 co-crystalized ligands and 3354 decoys randomly extracted from the DUD-E database | A subset of specifically selected 8918 purchasable compounds from the ZINC database | Ensemble of many pharmacophore models based on colchicine site-bound ligands structures was used in virtual screening which led to discovery of a potent tubulin-targeting cytotoxic agent |
| Zhang 2021 [32] | 1JFF | Discovery Studio 3.5 (Accelrys) | 467 inactive molecules from ZINC15 database and 33 known inhibitors with experimental activity | BioDiversity, 30,000 molecules | Large database filtered to focus on a subset that eventually led to discovery of two taxane-site targeting cytotoxic agents |
| Gallego-Yerga 2021 [84] | 1SA0 | Protein-ligand interaction fingerprints (PLIF) implemented in MOE (Chemical Computing Group Inc.) | No additional validation performed | A subset of 100, 000 compounds from ZINC15 database | Virtual screening campaign yielded a novel cytotoxic agent disrupting tubulin polymerization by binding at colchicine site |
| Elseginy 2022 [85] | 3E22, 3HKD, 3HKE, 3HKC, 1Z2B, and 1SA1 | Discovery Studio 2.5 (Accelrys) | 40 literature-mined tubulin inhibitors targeting the colchicine site, 2000 decoy molecules randomly selected from ChemDiv library | ChemDiv library, 700,000 molecules | A virtual screening campaign discovered an in vitro potent cytotoxic hit targeting the colchicine binding site |
| Screening Setup | Results | Reference | |||||
|---|---|---|---|---|---|---|---|
| Binding Site | Binding Site Definition | Docking Software | Screened Set | Hit No | Hit Rate, % | Best Compound’s Activity | |
| Virtual screening in succession to other computational methods | |||||||
| Colchicine | Extracted from 1SA0 as a 10 Å-wide cubic box around the center-of-mass of the native ligand | Glide SP | 25,146 virtual hits established by pharmacophore screening of CoCoCo database | 68 | 35% | Inhibition of tubulin polymerization at IC50 of 3 µM | Mangiatordi 2017 [18] |
| Colchicine | Extracted from 5H7O as 10 Å-wide cubic box around the center-of-mass of the native ligand | Glide SP, Glide XP | 30,327 virtual hits of pharmacophore screening of SPECS library | 8 | 20% | Anti-proliferative activity (IC50) against different cancer cell lines in range 6.14–15.06 µM | Guo 2020 [26] |
| Colchicine | Extracted from 6F7C (exact settings not specified) | MOE | 3135 virtual hits found by pharmacophore screening of SPECS library | 5 | 100% | 80% growth inhibition rate against five different cell lines | Zhou 2019 [83] |
| Taxane | Extracted from 1TVK as a grid box centered around the native ligand with each dimension a size of 5.8 Å | AutoDock 4.2 | 645 virtual hits yielded by similarity search in PubChem | 1 | 20% | Established hit got satisfactory predicted physiochemical properties; later work saw an analog compound synthesized and tested | Ayoub 2013 [16] |
| Taxane | Extracted from 1JFF as a sphere containing the residues within 11.5 Å from the ligand | AutoDock Vina | 1309 virtual hits established by a pharmacophore screening of the BioDiversity database | 11 | 22% | Anti-proliferative activity (IC50) against four cancer cell lines ranging from 10.31 µM to 21.04 µM | Zhang 2021 [32] |
| Gold | |||||||
| CDOCKER | |||||||
| Colchicine | Extracted from 1SA1 as all residues around the ligand at a 6.5 Å distance | SurFlex-Dock | 1739 virtual hits found by pharmacophore screening of the ChemDiv library | 1 | 1.78% | Tubulin polymerization inhibition IC50 value of 17.6 µM | Nagarajan 2015 [82] |
| Colchicine | Extracted from 4O2A as a sphere of 8 Å radius around the native ligand | GOLD | Around 3000 virtual hits procured by ligand-based virtual screening of six chemical libraries | 3 | 43% | IC50 of 83.61 µM in hepatotoxicity model | Federico 2020 [19] |
| Three databases: Chembridge Diverset EXP, Chembridge Diverset CL, and ZINC natural products | 4 | 66% | |||||
| Colchicine | Defined as a 20 Å-wide grid box around the centroid of the native ligand from the 1SA0 structure | MOE, BUDE, AutoDock 4.2 | 2746 virtual hits from a pharmacophore screening of a subset of ZINC15 library | 4 | 30% | Tubulin polymerization inhibition IC50 = 6.1 µM | Elseginy 2020 [85] |
| Taxane | Extracted from 1JFF as a 23 Å-wide box around the native ligand | AutoDock Vina | 1,601,806 compounds from the ChemDiv library | 1 | 5.8% | IC50 value against four cancer cells in range from 9.21 to 17.30 µM | Mao 2022 [90] |
| Colchicine | Extracted from 1SA0, 1SA1 (exact procedure not specified) | Glide SP | 1159 compounds from an in-house library | 6 | 35% | Tubulin polymerization inhibition at IC50 = 0.85 µM | Stefanski 2018 [27] |
| Virtual screening based on protein-ligand docking only | |||||||
| Peloruside | Extracted from 4O4J as a cubic grid of 20 Å in size | AutoDock 4.2 | 2000 virtual hits established after docking a 6 million ZINC subset with AutoDock Vina | 3 | 48% | Cell viability of HeLa cells decreased after 48 h by 60% at 100 µM | Zuniga-Bustos 2020 [91] |
| Colchicine | Extracted from 4O2B as all residues closer than 12 Å to the centroid of the native ligand | Glide SP, GOLD | 40,000 virtual hits obtained by high-throughput docking with Glide HTVS of IBScreen library | 2 | 13% | Tubulin polymerization inhibition IC50 = 23.5 µM | Liu 2022 [92] |
| Colchicine | Extracted from 4O2B as a cubic grid of 20 Å in size | AutoDock 4.2 | 212,449 compounds from the SPECS library | 2 | 5.5% | Tubulin polymerization inhibition activity with IC50 value of 1.68 μM | Liu 2019 [93] |
| Binding mode assessment | |||||||
| Colchicine | Extracted from 1SA0 as a 15 Å-wide cubic grid box centered on root point of native ligand | AutoDock 4.2 | An in-house library of 48 Schiff bases | 1 | – | Tubulin polymerization inhibition activity with IC50 value of 0.16 μM | Ameri 2018 [94] |
| Colchicine | Extracted from 4O2B as a sphere of 12 Å in diameter center on the native ligand | CDOCKER | A virtual hit from a ligand-based screening of the ChemDiv library | 1 | – | IC50 of 2.99 µM against CNE2 cancer cell line | Guo 2019 [17] |
| Colchicine | Extracted from 4O2B as a 30 Å-wide cubic grid box centered on root point of native ligand | AutoDock Vina | A single compound from an in-house designed library of colchicine site targeting ligands | 1 | – | IC50 = 0.6 µM in an anti-proliferative assay against the HeLa cancer cell line | Riu 2022 [95] |
| Colchicine | Extracted from 6Y6D as a 12 Å-wide grid box around the native ligand | Glide XP | In-house library of 9-arylimino noscapinoids | 3 | – | Anti-proliferative activity with IC50 of 10.8 µM against MCF-17 cancer cell line | Patel 2021 [96] |
| Colchicine | Extracted from 1SA0 as a 25 Å-wide box around the native ligand | AutoDock Vina | An in-house library of combretastatin A4 derivatives | 2 | – | Anti-proliferative activity with IC50 = 0.62 µM against HepG2 cancer cell line | Mustafa 2017 [97] |
| Taxane | Extracted from 1JFF and 1TUB a 30 Å-wide grid box around the native ligand | AutoDock 4.2 | Only a paclitaxel molecule was docked into tubulin mutants | 1 | – | Docking was used to provide rationale for paclitaxel resistance in mutant cancer cells | Tripathi 2016 [98] |
| Taxane | Extracted from 1TVK, 5MF4, 5LXT, and 3J6G as all residues within 6 Å distance from each native ligand | GOLD FRED | Only a lankacidin C molecule was docked into several conformations of taxane site | 1 | – | Ensemble docking was used to account for binding site flexibility and establish the binding mode of a recently discovered microtubules stabilizer targeting the taxane site | Ayoub 2019 [99] |
| Taxane | Extracted from 1JFF as a grid rectangle with a size of x = 30, y = 34, z = 26 centered on the native ligand | AutoDock 4 | A single hit with the best in vitro microtubule stabilizing properties | 1 | – | Binding to taxane suggested as a mechanism of action, promotion of tubulin polymerization by 76% at 50 µM | Chavez-Estrada 2020 [100] |
| Taxane | Extracted from 1JFF as a 21 Å-wide grid box centered on the native ligand | AutoDock 4 | Three compounds with the best in vitro anti-proliferative properties from a library of 32 marine natural and semisynthetic diterpenes | 3 | – | Interactions fingerprint analysis after docking prioritized the taxane site as the probable binding site for designed molecules with IC50 < 1 µM against three cancer cell lines | Forero 2021 [101] |
| Colchicine | Extracted from 1SA0 as a 21 Å-wide grid box centered on the native ligand | ||||||
| Vinca | Extracted from 4ZOL following an unspecified protocol | SurFlex-Dock | A known vinca-site ligand | 1 | – | Docking was used to guide the rational design of novel derivatives of tubulysin, which led to synthesis and validation of a hit with pronounced anti-proliferative properties attributed to binding at the vinca-site (IC50 = 9.4 nM against HeLa cell line) | Pandit 2021 [29] |
| Reference | PDB | Object of Study | MD Engine | Force field | Water Model | Time |
|---|---|---|---|---|---|---|
| Zhang 2019 [103] | 1Z2B | Docking refinement of DVB-α,β-tubulin complex | GROMACS 4.5 | SPC | 100 ns | |
| Majumdar 2019 [125] | 3HKB 3HKC | Comparison of the apo α,β-tubulin dimer and α,β-tubulin dimer bound to E7010 | NAMD 2.9 | Tubulin: CHARMM36 Ligand: CGenFF | TIP3P | 120 ns |
| Zhang 2021 [32] | 1JFF | Docking validation of ligand–tubulin complex for | GROMACS 2019.1 | Tubulin: Amber99sb-ildn Ligand: ACPYPE | SPC216 | 90 ns |
| Kumbhar 2021 [122] | 4O4J | Docking validation of PLA in complex with α,β-tubulin isotypes | GROMACS 5.0 | Tubulin: ff99SB-ildn Ligand: GAFF | TIP3P | 100 ns |
| Elhemely 2022 [116] | 4O2B | Docking of molecules at the colchicine site using an α,β-tubulin dimer. MD was used to study interactions and validate ligand persistence in binding site and SAR studies. | AMBER 19 | Tubulin: ff14SB Ligand: antechamber GAFF2 | TIP3P | 50 ns |
| Dash 2022 [119] | 1SA0 | Docking of molecules in the αβ-tubulin interface using a tubulin dimer. MD was used to study interactions, validate ligand persistence at the binding site, and calculate binding free energies. | AMBER 16 | Tubulin: ff14SB Ligand: GAFF | TIP3P | 100 ns |
| Hadizadeh 2022 [111] | 4O2B | Docking of molecules at the colchicine site. MD was used to study interactions and validate ligand persistence at the binding site. | NAMD 2.12 | Tubulin: CHARMM27 Ligand: provided by SwissParam | TIP3 | 100 ns |
| Mao 2022 [90] | 1JFF | Docking of molecules in the taxane site using a monomer of β-tubulin. MD was used to study interactions, validate ligand persistence at the binding site, and calculate binding free energies. | GROMACS 2019.1 | Tubulin: Amber99sb-ildn Ligand: ACPYPE | TIP3P | 80 ns |
| Neto 2022 [118] | 4O2B | Docking of chalcones in the colchicine site. MD was used to study interactions, validate ligand persistence at the binding site, and calculate binding free energies. | Discovery Studio software | implicit | 1000 ns | |
| Pragyandipta 2022 [126] | 6Y6D | Docking of molecules in the noscapinoids site. MD was used to study interactions, validate ligand persistence at the binding site, and calculate binding free energies. | GROMACS 2019.2 | Tubulin: GROMOS96 Ligand: ACPYPE | TIP3P | 100 ns |
| Yang 2022 [127] | 1JFF 4O4H | Study of wangzaozin as a binder for the taxane and laulimalide sites. | GROMACS 2019.1 | Tubulin: Amber99sb-ildn Ligand: ACPYPE | TIP3P | 90 ns |
| Boichuk 2022 [108] | 4O2B | Assess the position of the ligand at the colchicine binding site and determine key amino acid interactions using the EAPC-67-tubulin complex. | Desmond in Schrödinger suite 2021-2 | SPC | 100 ns | |
| Basu 2022 [121] | 1JFF 1TUB | Comparison of apo α,β-tubulin dimer, bound to taxol, and bound to Taxotere. | NAMD 2.11 | Tubulin: CHARMM36 Ligand: CGenFF | TIP3P | 200 ns |
| El-Mernissi 2022 [112] | 3E22 | 3E22-colchicine in complex with tubulin and two selected tubulin compound complexes to examine protein-ligand interactions. | Desmond Dynamics | OPLS | 50 ns | |
| Zhang 2022 [115] | 1JFF | Docking validation of hits bound to the taxane site | GROMACS 2019.1 | Tubulin: Amber99sb-ildn | SPC216 | 90 ns |
| Zhao 2022 [115] | 4O2B | Docking validation of styrylquinoline tubulin inhibitors | AMBER16 | Tubulin: Amber ff99SB Ligand: GAFF | TIP3P | 100 ns |
| Radha 2022 [123] | 6Y6D | Docking validation of shikonin as a tubulin inhibitor | GROMACS 2019.2 | Tubulin: Amber ff99SB Ligand: GAFF | TIP3P | 100 ns |
| Rai 2022 [107] | MD used for the analysis of the Interactions between eribulin and different tubulin isotypes | AMBER 12 | Tubulin: Amber ff99SB Ligand: Antechamber tool | implicit | 60 ns |
References
- Pellegrini, L.; Wetzel, A.; Granno, S.; Heaton, G.; Harvey, K. Back to the tubule: Microtubule dynamics in Parkinson’s disease. Cell. Mol. Life Sci. 2017, 74, 409–434. [Google Scholar] [CrossRef]
- Čermák, V.; Dostál, V.; Jelínek, M.; Libusová, L.; Kovář, J.; Rösel, D.; Brábek, J. Microtubule-targeting agents and their impact on cancer treatment. Eur. J. Cell Biol. 2020, 99, 151075. [Google Scholar] [CrossRef] [PubMed]
- Bracey, K.M.; Ho, K.H.; Yampolsky, D.; Gu, G.; Kaverina, I.; Holmes, W.R. Microtubules Regulate Localization and Availability of Insulin Granules in Pancreatic Beta Cells. Biophys. J. 2020, 118, 193–206. [Google Scholar] [CrossRef]
- Dubey, J.; Ratnakaran, N.; Koushika, S.P. Neurodegeneration and microtubule dynamics: Death by a thousand cuts. Front. Cell. Neurosci. 2015, 9, 343. [Google Scholar] [CrossRef]
- Mitchison, T.; Kirschner, M. Dynamic instability of microtubule growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef]
- Kollman, J.M.; Merdes, A.; Mourey, L.; Agard, D.A. Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol. 2011, 12, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Teixido-Travesa, N.; Roig, J.; Luders, J. The where, when and how of microtubule nucleation-one ring to rule them all. J. Cell Sci. 2012, 125, 4445–4456. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wurtz, M.; Zupa, E.; Pfeffer, S.; Schiebel, E. Microtubule nucleation: The waltz between gamma-tubulin ring complex and associated proteins. Curr. Opin. Cell Biol. 2021, 68, 124–131. [Google Scholar] [CrossRef]
- Brouhard, G.J.; Rice, L.M. Microtubule dynamics: An interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 2018, 19, 451–463. [Google Scholar] [CrossRef]
- Roostalu, J.; Thomas, C.; Cade, N.I.; Kunzelmann, S.; Taylor, I.A.; Surrey, T. The speed of GTP hydrolysis determines GTP cap size and controls microtubule stability. Elife 2020, 9, e51992. [Google Scholar] [CrossRef]
- Mühlethaler, T.; Gioia, D.; Prota, A.E.; Sharpe, M.E.; Cavalli, A.; Steinmetz, M.O. Comprehensive Analysis of Binding Sites in Tubulin. Angew. Chem. Int. Ed. Engl. 2021, 60, 13331–13342. [Google Scholar] [CrossRef] [PubMed]
- Muhlethaler, T.; Milanos, L.; Ortega, J.A.; Blum, T.B.; Gioia, D.; Roy, B.; Prota, A.E.; Cavalli, A.; Steinmetz, M.O. Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action. Angew. Chem. Int. Ed. Engl. 2022, 61, e202204052. [Google Scholar] [CrossRef] [PubMed]
- Marzaro, G.; Chilin, A. QSAR and 3D-QSAR models in the field of tubulin inhibitors as anticancer agents. Curr. Top. Med. Chem. 2014, 14, 2253–2262. [Google Scholar] [CrossRef]
- Johnson, M.A.; Maggiora, G.M. Concepts and Applications of Molecular Similarity; Wiley: Hoboken, NJ, USA, 1990. [Google Scholar]
- Horvath, D.; Koch, C.; Schneider, G.; Marcou, G.; Varnek, A. Local neighborhood behavior in a combinatorial library context. J. Comput. Aided. Mol. Des. 2011, 25, 237–252. [Google Scholar] [CrossRef]
- Ayoub, A.T.; Klobukowski, M.; Tuszynski, J. Similarity-based virtual screening for microtubule stabilizers reveals novel antimitotic scaffold. J. Mol. Graph. Model. 2013, 44, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Luo, Y.; Zhai, S.; Jiang, Z.; Zhao, C.; Xu, J.; Wang, L. Discovery, biological evaluation, structure-activity relationships and mechanism of action of pyrazolo[3,4-b]pyridin-6-one derivatives as a new class of anticancer agents. Org. Biomol. Chem. 2019, 17, 6201–6214. [Google Scholar] [CrossRef] [PubMed]
- Mangiatordi, G.F.; Trisciuzzi, D.; Alberga, D.; Denora, N.; Iacobazzi, R.M.; Gadaleta, D.; Catto, M.; Nicolotti, O. Novel chemotypes targeting tubulin at the colchicine binding site and unbiasing P-glycoprotein. Eur. J. Med. Chem. 2017, 139, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Federico, L.B.; Silva, G.M.; de Fraga Dias, A.; Figueiro, F.; Battastini, A.M.O.; Dos Santos, C.B.R.; Costa, L.T.; Rosa, J.M.C.; de Paula da Silva, C.H.T. Identification of novel alphabeta-tubulin modulators with antiproliferative activity directed to cancer therapy using ligand and structure-based virtual screening. Int. J. Biol. Macromol. 2020, 165, 3040–3050. [Google Scholar] [CrossRef]
- Lo, Y.C.; Senese, S.; Li, C.M.; Hu, Q.; Huang, Y.; Damoiseaux, R.; Torres, J.Z. Large-scale chemical similarity networks for target profiling of compounds identified in cell-based chemical screens. PLoS Comput. Biol. 2015, 11, e1004153. [Google Scholar] [CrossRef]
- Lo, Y.C.; Senese, S.; Damoiseaux, R.; Torres, J.Z. 3D Chemical Similarity Networks for Structure-Based Target Prediction and Scaffold Hopping. ACS Chem. Biol. 2016, 11, 2244–2253. [Google Scholar] [CrossRef]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.; Oprea, T.I.; Baskin, I.I.; Varnek, A.; Roitberg, A.; et al. QSAR without borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef]
- Tropsha, A.; Gramatica, P.; Gombar, V.K. The importance of being earnest: Validation is the absolute essential for successful application and interpretation of QSPR models. Qsar Comb. Sci. 2003, 22, 69–77. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, R.; Amin, S.A.; Adhikari, N.; Ghorai, S.; Jha, T.; Gayen, S. Identification of molecular fingerprints of phenylindole derivatives as cytotoxic agents: A multi-QSAR approach. Struct. Chem. 2018, 29, 1095–1107. [Google Scholar] [CrossRef]
- Guo, Q.; Zhang, H.; Deng, Y.; Zhai, S.; Jiang, Z.; Zhu, D.; Wang, L. Ligand- and structural-based discovery of potential small molecules that target the colchicine site of tubulin for cancer treatment. Eur. J. Med. Chem. 2020, 196, 112328. [Google Scholar] [CrossRef]
- Stefanski, T.; Mikstacka, R.; Kurczab, R.; Dutkiewicz, Z.; Kucinska, M.; Murias, M.; Zielinska-Przyjemska, M.; Cichocki, M.; Teubert, A.; Kaczmarek, M.; et al. Design, synthesis, and biological evaluation of novel combretastatin A-4 thio derivatives as microtubule targeting agents. Eur. J. Med. Chem. 2018, 144, 797–816. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.P.; Cheng, L.P.; Wang, T.C.; Pang, W.; Wu, F.H.; Huang, J.W. Molecular modeling study, synthesis and biological evaluation of combretastatin A-4 analogues as anticancer agents and tubulin inhibitors. Medchemcomm 2018, 9, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Pandit, A.; Yadav, K.; Reddy, R.B.; Sengupta, S.; Sharma, R.; Chelvam, V. Structure activity relationships (SAR) study to design and synthesize new tubulin inhibitors with enhanced anti-tubulin activity: In silico and in vitro analysis. J. Mol. Struct. 2021, 1223, 129204. [Google Scholar] [CrossRef]
- Giordano, D.; Biancaniello, C.; Argenio, M.A.; Facchiano, A. Drug Design by Pharmacophore and Virtual Screening Approach. Pharmaceuticals 2022, 15, 646. [Google Scholar] [CrossRef]
- Seidel, T.; Wieder, O.; Garon, A.; Langer, T. Applications of the Pharmacophore Concept in Natural Product inspired Drug Design. Mol. Inform. 2020, 39, e2000059. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, J.; Yang, Y.L.; Liu, C.T.; Shen, C.; Zhang, H.R.; Xie, H.Z.; Ding, L. Discovery of novel tubulin inhibitors targeting taxanes site by virtual screening, molecular dynamic simulation, and biological evaluation. J. Cell. Biochem. 2021, 122, 1609–1624. [Google Scholar] [CrossRef] [PubMed]
- Lone, M.Y.; Athar, M.; Manhas, A.; Jha, P.C.; Bhatt, S.; Shah, A. In Silico Exploration of Vinca Domain Tubulin Inhibitors: A Combination of 3D-QSAR-Based Pharmacophore Modeling, Docking and Molecular Dynamics Simulations. ChemistrySelect 2017, 2, 10848–10853. [Google Scholar] [CrossRef]
- Niu, M.M.; Qin, J.Y.; Tian, C.P.; Yan, X.F.; Dong, F.G.; Cheng, Z.Q.; Fida, G.; Yang, M.; Chen, H.Y.; Gu, Y.Q. Tubulin inhibitors: Pharmacophore modeling, virtual screening and molecular docking. Acta Pharmacol. Sin. 2014, 35, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Urvoas, A.; Consolati, T.; Cantos-Fernandes, S.; Aumont-Nicaise, M.; Valerio-Lepiniec, M.; Surrey, T.; Minard, P.; Gigant, B. Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit. Structure 2019, 27, 497–506.e494. [Google Scholar] [CrossRef]
- Curmi, P.A.; Andersen, S.S.; Lachkar, S.; Gavet, O.; Karsenti, E.; Knossow, M.; Sobel, A. The stathmin/tubulin interaction in vitro. J. Biol. Chem. 1997, 272, 25029–25036. [Google Scholar] [CrossRef]
- Steinmetz, M.O.; Kammerer, R.A.; Jahnke, W.; Goldie, K.N.; Lustig, A.; van Oostrum, J. Op18/stathmin caps a kinked protofilament-like tubulin tetramer. EMBO J. 2000, 19, 572–580. [Google Scholar] [CrossRef]
- Nogales, E.; Wolf, S.G.; Downing, K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature 1998, 391, 199–203. [Google Scholar] [CrossRef]
- Gigant, B.; Curmi, P.A.; Martin-Barbey, C.; Charbaut, E.; Lachkar, S.; Lebeau, L.; Siavoshian, S.; Sobel, A.; Knossow, M. The 4 angstrom X-ray structure of a tubulin: Stathmin-like domain complex. Cell 2000, 102, 809–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravelli, R.B.G.; Gigant, B.; Curmi, P.A.; Jourdain, I.; Lachkar, S.; Sobel, A.; Knossow, M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004, 428, 198–202. [Google Scholar] [CrossRef]
- Prota, A.E.; Magiera, M.M.; Kuijpers, M.; Bargsten, K.; Frey, D.; Wieser, M.; Jaussi, R.; Hoogenraad, C.C.; Kammerer, R.A.; Janke, C.; et al. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 2013, 200, 259–270. [Google Scholar] [CrossRef]
- Prota, A.E.; Bargsten, K.; Zurwerra, D.; Field, J.J.; Díaz, J.F.; Altmann, K.-H.; Steinmetz, M.O. Molecular Mechanism of Action of Microtubule-Stabilizing Anticancer Agents. Science 2013, 339, 587–590. [Google Scholar] [CrossRef]
- Pecqueur, L.; Duellberg, C.; Dreier, B.; Jiang, Q.; Wang, C.; Pluckthun, A.; Surrey, T.; Gigant, B.; Knossow, M. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc. Natl. Acad. Sci. USA 2012, 109, 12011–12016. [Google Scholar] [CrossRef]
- La Sala, G.; Olieric, N.; Sharma, A.; Viti, F.; Perez, F.D.B.; Huang, L.; Tonra, J.R.; Lloyd, G.K.; Decherchi, S.; Diaz, J.F.; et al. Structure, Thermodynamics, and Kinetics of Plinabulin Binding to Two Tubulin Isotypes. Chem 2019, 5, 2969–2986. [Google Scholar] [CrossRef]
- Ayaz, P.; Ye, X.; Huddleston, P.; Brautigam, C.A.; Rice, L.M. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 2012, 337, 857–860. [Google Scholar] [CrossRef]
- Campanacci, V.; Urvoas, A.; Ammar Khodja, L.; Aumont-Nicaise, M.; Noiray, M.; Lachkar, S.; Curmi, P.A.; Minard, P.; Gigant, B. Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors. Proc. Natl. Acad. Sci. USA 2022, 119, e2120098119. [Google Scholar] [CrossRef]
- Sharma, A.; Aher, A.; Dynes, N.J.; Frey, D.; Katrukha, E.A.; Jaussi, R.; Grigoriev, I.; Croisier, M.; Kammerer, R.A.; Akhmanova, A.; et al. Centriolar CPAP/SAS-4 Imparts Slow Processive Microtubule Growth. Dev. Cell 2016, 37, 362–376. [Google Scholar] [CrossRef]
- Zheng, X.; Ramani, A.; Soni, K.; Gottardo, M.; Zheng, S.; Ming Gooi, L.; Li, W.; Feng, S.; Mariappan, A.; Wason, A.; et al. Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat. Commun. 2016, 7, 11874. [Google Scholar] [CrossRef]
- Steinmetz, M.O.; Prota, A.E. Microtubule-Targeting Agents: Strategies To Hijack the Cytoskeleton. Trends Cell Biol. 2018, 28, 776–792. [Google Scholar] [CrossRef]
- Elie-Caille, C.; Severin, F.; Helenius, J.; Howard, J.; Muller, D.J.; Hyman, A.A. Straight GDP-tubulin protofilaments form in the presence of taxol. Curr. Biol. 2007, 17, 1765–1770. [Google Scholar] [CrossRef] [PubMed]
- Alushin, G.M.; Lander, G.C.; Kellogg, E.H.; Zhang, R.; Baker, D.; Nogales, E. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell 2014, 157, 1117–1129. [Google Scholar] [CrossRef]
- Lowe, J.; Li, H.; Downing, K.H.; Nogales, E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J. Mol. Biol. 2001, 313, 1045–1057. [Google Scholar] [CrossRef]
- Kellogg, E.H.; Hejab, N.M.A.; Howes, S.; Northcote, P.; Miller, J.H.; Diaz, J.F.; Downing, K.H.; Nogales, E. Insights into the Distinct Mechanisms of Action of Taxane and Non-Taxane Microtubule Stabilizers from Cryo-EM Structures. J. Mol. Biol. 2017, 429, 633–646. [Google Scholar] [CrossRef]
- Prota, A.E.; Bargsten, K.; Northcote, P.T.; Marsh, M.; Altmann, K.H.; Miller, J.H.; Diaz, J.F.; Steinmetz, M.O. Structural basis of microtubule stabilization by laulimalide and peloruside A. Angew. Chem. Int. Ed. Engl. 2014, 53, 1621–1625. [Google Scholar] [CrossRef]
- Dorleans, A.; Gigant, B.; Ravelli, R.B.G.; Mailliet, P.; Mikol, V.; Knossow, M. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc. Natl. Acad. Sci. USA 2009, 106, 13775–13779. [Google Scholar] [CrossRef] [PubMed]
- Gigant, B.; Wang, C.; Ravelli, R.B.; Roussi, F.; Steinmetz, M.O.; Curmi, P.A.; Sobel, A.; Knossow, M. Structural basis for the regulation of tubulin by vinblastine. Nature 2005, 435, 519–522. [Google Scholar] [CrossRef]
- Cormier, A.; Marchand, M.; Ravelli, R.B.; Knossow, M.; Gigant, B. Structural insight into the inhibition of tubulin by vinca domain peptide ligands. EMBO Rep. 2008, 9, 1101–1106. [Google Scholar] [CrossRef]
- Maderna, A.; Doroski, M.; Subramanyam, C.; Porte, A.; Leverett, C.A.; Vetelino, B.C.; Chen, Z.; Risley, H.; Parris, K.; Pandit, J.; et al. Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications. J. Med. Chem. 2014, 57, 10527–10543. [Google Scholar] [CrossRef] [PubMed]
- Prota, A.E.; Bargsten, K.; Diaz, J.F.; Marsh, M.; Cuevas, C.; Liniger, M.; Neuhaus, C.; Andreu, J.M.; Altmann, K.H.; Steinmetz, M.O. A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs. Proc. Natl. Acad. Sci. USA 2014, 111, 13817–13821. [Google Scholar] [CrossRef] [Green Version]
- Prota, A.E.; Setter, J.; Waight, A.B.; Bargsten, K.; Murga, J.; Diaz, J.F.; Steinmetz, M.O. Pironetin Binds Covalently to alphaCys316 and Perturbs a Major Loop and Helix of alpha-Tubulin to Inhibit Microtubule Formation. J. Mol. Biol. 2016, 428, 2981–2988. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Y.; Wang, T.; Jiang, J.; Botting, C.H.; Liu, H.; Chen, Q.; Yang, J.; Naismith, J.H.; Zhu, X.; et al. Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule. Nat. Commun. 2016, 7, 12103. [Google Scholar] [CrossRef]
- Matthew, S.; Chen, Q.Y.; Ratnayake, R.; Fermaintt, C.S.; Lucena-Agell, D.; Bonato, F.; Prota, A.E.; Lim, S.T.; Wang, X.; Diaz, J.F.; et al. Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site. Proc. Natl. Acad. Sci. USA 2021, 118, e2021847118. [Google Scholar] [CrossRef]
- Prota, A.E.; Danel, F.; Bachmann, F.; Bargsten, K.; Buey, R.M.; Pohlmann, J.; Reinelt, S.; Lane, H.; Steinmetz, M.O. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J. Mol. Biol. 2014, 426, 1848–1860. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.A.; Prota, A.E.; Rodríguez-Salarichs, J.; Bennani, Y.L.; Jiménez-Barbero, J.; Bargsten, K.; Canales, Á.; Steinmetz, M.O.; Díaz, J.F. Structural Basis of Noscapine Activation for Tubulin Binding. J. Med. Chem. 2020, 63, 8495–8501. [Google Scholar] [CrossRef] [PubMed]
- Doodhi, H.; Prota, A.E.; Rodriguez-Garcia, R.; Xiao, H.; Custar, D.W.; Bargsten, K.; Katrukha, E.A.; Hilbert, M.; Hua, S.; Jiang, K.; et al. Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes. Curr. Biol. 2016, 26, 1713–1721. [Google Scholar] [CrossRef]
- Weinert, T.; Olieric, N.; Cheng, R.; Brunle, S.; James, D.; Ozerov, D.; Gashi, D.; Vera, L.; Marsh, M.; Jaeger, K.; et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 2017, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Nawrotek, A.; Knossow, M.; Gigant, B. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J. Mol. Biol. 2011, 412, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Prota, A.E.; Bargsten, K.; Redondo-Horcajo, M.; Smith, A.B., III; Yang, C.H.; McDaid, H.M.; Paterson, I.; Horwitz, S.B.; Fernando Diaz, J.; Steinmetz, M.O. Structural Basis of Microtubule Stabilization by Discodermolide. Chembiochem 2017, 18, 905–909. [Google Scholar] [CrossRef]
- Guo, B.; Rodriguez-Gabin, A.; Prota, A.E.; Muhlethaler, T.; Zhang, N.; Ye, K.; Steinmetz, M.O.; Horwitz, S.B.; Smith, A.B., III; McDaid, H.M. Structural Refinement of the Tubulin Ligand (+)-Discodermolide to Attenuate Chemotherapy-Mediated Senescence. Mol. Pharmacol. 2020, 98, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Menchon, G.; Prota, A.E.; Lucena-Agell, D.; Bucher, P.; Jansen, R.; Irschik, H.; Muller, R.; Paterson, I.; Diaz, J.F.; Altmann, K.H.; et al. A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin. Nat. Commun. 2018, 9, 2106. [Google Scholar] [CrossRef]
- Gao, L.; Meiring, J.C.M.; Kraus, Y.; Wranik, M.; Weinert, T.; Pritzl, S.D.; Bingham, R.; Ntouliou, E.; Jansen, K.I.; Olieric, N.; et al. A Robust, GFP-Orthogonal Photoswitchable Inhibitor Scaffold Extends Optical Control over the Microtubule Cytoskeleton. Cell Chem. Biol. 2021, 28, 228–241.e226. [Google Scholar] [CrossRef] [PubMed]
- De la Roche, N.M.; Muhlethaler, T.; Di Martino, R.M.C.; Ortega, J.A.; Gioia, D.; Roy, B.; Prota, A.E.; Steinmetz, M.O.; Cavalli, A. Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents. Eur. J. Med. Chem. 2022, 241, 114614. [Google Scholar] [CrossRef] [PubMed]
- Bohnacker, T.; Prota, A.E.; Beaufils, F.; Burke, J.E.; Melone, A.; Inglis, A.J.; Rageot, D.; Sele, A.M.; Cmiljanovic, V.; Cmiljanovic, N.; et al. Deconvolution of Buparlisib’s mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention. Nat. Commun. 2017, 8, 14683. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Hilken Nee Thomopoulou, P.; Frias, C.; Hopff, S.M.; Varela, P.; Wilke, N.; Mariappan, A.; Neudorfl, J.M.; Fedorov, A.Y.; Gopalakrishnan, J.; et al. B-nor-methylene Colchicinoid PT-100 Selectively Induces Apoptosis in Multidrug-Resistant Human Cancer Cells via an Intrinsic Pathway in a Caspase-Independent Manner. ACS Omega 2022, 7, 2591–2603. [Google Scholar] [CrossRef] [PubMed]
- Waight, A.B.; Bargsten, K.; Doronina, S.; Steinmetz, M.O.; Sussman, D.; Prota, A.E. Structural Basis of Microtubule Destabilization by Potent Auristatin Anti-Mitotics. PLoS ONE 2016, 11, e0160890. [Google Scholar] [CrossRef]
- Debs, G.E.; Cha, M.; Liu, X.; Huehn, A.R.; Sindelar, C.V. Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 16976–16984. [Google Scholar] [CrossRef]
- LaFrance, B.J.; Roostalu, J.; Henkin, G.; Greber, B.J.; Zhang, R.; Normanno, D.; McCollum, C.O.; Surrey, T.; Nogales, E. Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules. Proc. Natl. Acad. Sci. USA 2022, 119, e2114994119. [Google Scholar] [CrossRef]
- Castro-Alvarez, A.; Pineda, O.; Vilarrasa, J. Further Insight into the Interactions of the Cytotoxic Macrolides Laulimalide and Peloruside A with Their Common Binding Site. ACS Omega 2018, 3, 1770–1782. [Google Scholar] [CrossRef]
- Gaurav, A.; Gautam, V. Structure-based three-dimensional pharmacophores as an alternative to traditional methodologies. J. Recept. Ligand Channel Res. 2014, 2014, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, S.; Choi, M.J.; Cho, Y.S.; Min, S.J.; Keum, G.; Kim, S.J.; Lee, C.S.; Pae, A.N. Tubulin inhibitor identification by bioactive conformation alignment pharmacophore-guided virtual screening. Chem. Biol. Drug Des. 2015, 86, 998–1016. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Di, B.; Niu, M.M. Structure-Based Pharmacophore Design and Virtual Screening for Novel Tubulin Inhibitors with Potential Anticancer Activity. Molecules 2019, 24, 3181. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Yerga, L.; Ochoa, R.; Lans, I.; Pena-Varas, C.; Alegria-Arcos, M.; Cossio, P.; Ramirez, D.; Pelaez, R. Application of ensemble pharmacophore-based virtual screening to the discovery of novel antimitotic tubulin inhibitors. Comput. Struct. Biotechnol. J. 2021, 19, 4360–4372. [Google Scholar] [CrossRef]
- Elseginy, S.A.; Oliveira, A.S.F.; Shoemark, D.K.; Sessions, R.B. Identification and validation of novel microtubule suppressors with an imidazopyridine scaffold through structure-based virtual screening and docking. RSC Med. Chem. 2022, 13, 929–943. [Google Scholar] [CrossRef]
- Sulimov, V.B.; Kutov, D.C.; Sulimov, A.V. Advances in Docking. Curr. Med. Chem. 2019, 26, 7555–7580. [Google Scholar] [CrossRef]
- Halperin, I.; Ma, B.; Wolfson, H.; Nussinov, R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins 2002, 47, 409–443. [Google Scholar] [CrossRef]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, G.E.M.; Govender, T.; Naicker, T.; Kruger, H.G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. 2021, 224, 113705. [Google Scholar] [CrossRef]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- Mao, J.; Luo, Q.Q.; Zhang, H.R.; Zheng, X.H.; Shen, C.; Qi, H.Z.; Hu, M.L.; Zhang, H. Discovery of microtubule stabilizers with novel scaffold structures based on virtual screening, biological evaluation, and molecular dynamics simulation. Chem. Biol. Interact. 2022, 352, 109784. [Google Scholar] [CrossRef]
- Zuniga-Bustos, M.; Vasquez, P.A.; Jana, G.A.; Guzman, J.L.; Alderete, J.B.; Jimenez, V.A. Mechanism-Based Rational Discovery and In Vitro Evaluation of Novel Microtubule Stabilizing Agents with Non-Taxol-Competitive Activity. J. Chem. Inf. Model. 2020, 60, 3204–3213. [Google Scholar] [CrossRef]
- Liu, W.; Jia, H.; Guan, M.; Cui, M.; Lan, Z.; He, Y.; Guo, Z.; Jiang, R.; Dong, G.; Wang, S. Discovery of novel tubulin inhibitors targeting the colchicine binding site via virtual screening, structural optimization and antitumor evaluation. Bioorganic Chem. 2022, 118, 105486. [Google Scholar] [CrossRef]
- Liu, G.; Jiao, Y.; Huang, C.; Chang, P. Identification of novel and potent small-molecule inhibitors of tubulin with antitumor activities by virtual screening and biological evaluations. J. Comput. Aided. Mol. Des. 2019, 33, 659–664. [Google Scholar] [CrossRef]
- Ameri, A.; Khodarahmi, G.; Forootanfar, H.; Hassanzadeh, F.; Hakimelahi, G.H. Hybrid Pharmacophore Design, Molecular Docking, Synthesis, and Biological Evaluation of Novel Aldimine-Type Schiff Base Derivatives as Tubulin Polymerization Inhibitor. Chem. Biodivers. 2018, 15, e1700518. [Google Scholar] [CrossRef]
- Riu, F.; Ibba, R.; Zoroddu, S.; Sestito, S.; Lai, M.; Piras, S.; Sanna, L.; Bordoni, V.; Bagella, L.; Carta, A. Design, synthesis, and biological screening of a series of 4’-fluoro-benzotriazole-acrylonitrile derivatives as microtubule-destabilising agents (MDAs). J. Enzyme. Inhib. Med. Chem. 2022, 37, 2223–2240. [Google Scholar] [CrossRef]
- Patel, A.K.; Meher, R.K.; Nagireddy, P.K.; Pragyandipta, P.; Pedapati, R.K.; Kantevari, S.; Naik, P.K. 9-Arylimino noscapinoids as potent tubulin binding anticancer agent: Chemical synthesis and cellular evaluation against breast tumour cells. SAR QSAR Environ. Res. 2021, 32, 269–291. [Google Scholar] [CrossRef]
- Mustafa, M.; Abdelhamid, D.; Abdelhafez, E.M.N.; Ibrahim, M.A.A.; Gamal-Eldeen, A.M.; Aly, O.M. Synthesis, antiproliferative, anti-tubulin activity, and docking study of new 1,2,4-triazoles as potential combretastatin analogues. Eur. J. Med. Chem. 2017, 141, 293–305. [Google Scholar] [CrossRef]
- Tripathi, S.; Srivastava, G.; Sharma, A. Molecular dynamics simulation and free energy landscape methods in probing L215H, L217R and L225M betaI-tubulin mutations causing paclitaxel resistance in cancer cells. Biochem. Biophys. Res. Commun. 2016, 476, 273–279. [Google Scholar] [CrossRef]
- Ayoub, A.T.; Elrefaiy, M.A.; Arakawa, K. Computational Prediction of the Mode of Binding of Antitumor Lankacidin C to Tubulin. ACS Omega 2019, 4, 4461–4471. [Google Scholar] [CrossRef]
- Chávez-Estrada, E.J.; Cerda-García-Rojas, C.M.; Román-Marín, L.U.; Hernández-Hernández, J.D.; Joseph-Nathan, P. Synthesis, molecular docking, and saturation-transfer difference NMR spectroscopy of longipinane derivatives as novel microtubule stabilizers. J. Mol. Struct. 2020, 1218, 128519. [Google Scholar] [CrossRef]
- Forero, A.M.; Castellanos, L.; Sandoval-Hernandez, A.G.; Magalhaes, A.; Tinoco, L.W.; Lopez-Vallejo, F.; Ramos, F.A. Integration of NMR studies, computational predictions, and in vitro assays in the search of marine diterpenes with antitumor activity. Chem. Biol. Drug Des. 2021, 98, 507–521. [Google Scholar] [CrossRef]
- Ngo, S.T.; Vu, K.B.; Bui, L.M.; Vu, V.V. Effective Estimation of Ligand-Binding Affinity Using Biased Sampling Method. ACS Omega 2019, 4, 3887–3893. [Google Scholar] [CrossRef]
- Zhang, Z.; Lu, C.; Wang, P.; Li, A.; Zhang, H.; Xu, S. Structural Basis and Mechanism for Vindoline Dimers Interacting with α,β-Tubulin. ACS Omega 2019, 4, 11938–11948. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, Z.; Li, A.; Zhang, Z.; Xu, S. Double-sides sticking mechanism of vinblastine interacting with α,β-tubulin to get activity against cancer cells. J. Biomol. Struct. Dyn. 2018, 37, 4080–4091. [Google Scholar] [CrossRef]
- Mane, J.Y.; Semenchenko, V.; Perez-Pineiro, R.; Winter, P.; Wishart, D.; Tuszynski, J.A. Experimental and computational study of the interaction of novel colchicinoids with a recombinant human alphaI/betaI-tubulin heterodimer. Chem. Biol. Drug Des. 2013, 82, 60–70. [Google Scholar] [CrossRef]
- Izrailev, S.; Stepaniants, S.; Balsera, M.; Oono, Y.; Schulten, K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997, 72, 1568–1581. [Google Scholar] [CrossRef]
- Rai, K.; Kumbhar, B.V.; Panda, D.; Kunwar, A. Computational study of interactions of anti-cancer drug eribulin with human tubulin isotypes. Phys. Chem. Chem. Phys. 2022, 24, 16694–16700. [Google Scholar] [CrossRef]
- Boichuk, S.; Syuzov, K.; Bikinieva, F.; Galembikova, A.; Zykova, S.; Gankova, K.; Igidov, S.; Igidov, N. Computational-Based Discovery of the Anti-Cancer Activities of Pyrrole-Based Compounds Targeting the Colchicine-Binding Site of Tubulin. Molecules 2022, 27, 2873. [Google Scholar] [CrossRef]
- Fusani, L.; Palmer, D.S.; Somers, D.O.; Wall, I.D. Exploring Ligand Stability in Protein Crystal Structures Using Binding Pose Metadynamics. J. Chem. Inf. Model. 2020, 60, 1528–1539. [Google Scholar] [CrossRef]
- Gaspari, R.; Prota, A.E.; Bargsten, K.; Cavalli, A.; Steinmetz, M.O. Structural Basis of cis- and trans-Combretastatin Binding to Tubulin. Chem 2017, 2, 102–113. [Google Scholar] [CrossRef]
- Hadizadeh, F.; Ghodsi, R.; Mirzaei, S.; Sahebkar, A. In Silico Exploration of Novel Tubulin Inhibitors: A Combination of Docking and Molecular Dynamics Simulations, Pharmacophore Modeling, and Virtual Screening. Comput. Math. Methods Med. 2022, 2022, 4004068. [Google Scholar] [CrossRef]
- El-Mernissi, R.; El Khatabi, K.; Khaldan, A.; ElMchichi, L.; Shahinozzaman, M.; Ajana, M.A.; Lakhlifi, T.; Bouachrine, M. 2-Oxoquinoline Arylaminothiazole Derivatives in Identifying Novel Potential Anticancer Agents by Applying 3D-QSAR, Docking, and Molecular Dynamics Simulation Studies. J. Mex. Chem. Soc. 2021, 66. [Google Scholar] [CrossRef]
- Gonzalez-Aleman, R.; Hernandez-Castillo, D.; Rodriguez-Serradet, A.; Caballero, J.; Hernandez-Rodriguez, E.W.; Montero-Cabrera, L. BitClust: Fast Geometrical Clustering of Long Molecular Dynamics Simulations. J. Chem. Inf. Model. 2020, 60, 444–448. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Zhang, H.; Qi, H.Z.; Mao, J.; Zhang, H.R.; Luo, Q.Q.; Hu, M.L.; Shen, C.; Ding, L. Discovery of novel microtubule stabilizers targeting taxane binding site by applying molecular docking, molecular dynamics simulation, and anticancer activity testing. Bioorganic Chem. 2022, 122, 105722. [Google Scholar] [CrossRef]
- Elhemely, M.A.; Belgath, A.A.; El-Sayed, S.; Burusco, K.K.; Kadirvel, M.; Tirella, A.; Finegan, K.; Bryce, R.A.; Stratford, I.J.; Freeman, S. SAR of Novel 3-Arylisoquinolinones: Meta-Substitution on the Aryl Ring Dramatically Enhances Antiproliferative Activity through Binding to Microtubules. J. Med. Chem. 2022, 65, 4783–4797. [Google Scholar] [CrossRef]
- Stroylov, V.S.; Svitanko, I.V.; Maksimenko, A.S.; Kislyi, V.P.; Semenova, M.N.; Semenov, V.V. Computational modeling and target synthesis of monomethoxy-substituted o-diphenylisoxazoles with unexpectedly high antimitotic microtubule destabilizing activity. Bioorganic Med. Chem. Let.t 2020, 30, 127608. [Google Scholar] [CrossRef]
- Neto, R.A.M.; Santos, C.B.R.; Henriques, S.V.C.; Machado, L.O.; Cruz, J.N.; da Silva, C.; Federico, L.B.; Oliveira, E.H.C.; de Souza, M.P.C.; da Silva, P.N.B.; et al. Novel chalcones derivatives with potential antineoplastic activity investigated by docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2022, 40, 2204–2216. [Google Scholar] [CrossRef]
- Dash, S.G.; Naik, P.K. 10. 9-VINYL PHENYL NOSCAPINE AS POTENTIAL TUBULIN BINDING ANTICANCER AGENT. Biotechnology 2022, 102, 102. [Google Scholar]
- Zhao, X.; Zhang, R.; Yu, X.; Yu, N.; Shi, Y.; Shu, M.; Shen, Y. Discovery of novel tubulin polymerization inhibitors by utilizing 3D-QSAR, molecular docking and molecular dynamics simulation. New J. Chem. 2022, 46, 16426–16435. [Google Scholar] [CrossRef]
- Basu, D.; Majumdar, S.; Mandal, N.; Dastidar, S.G. Mechanisms of influence of the microtubule over-stabilizing ligands on the structure and intrinsic dynamics of α,β-Tubulin. Comput. Biol. Chem. 2022, 96, 107617. [Google Scholar] [CrossRef]
- Kumbhar, B.V.; Bhandare, V.V. Exploring the interaction of Peloruside-A with drug resistant alphabetaII and alphabetaIII tubulin isotypes in human ovarian carcinoma using a molecular modeling approach. J. Biomol. Struct. Dyn. 2021, 39, 1990–2002. [Google Scholar] [CrossRef] [PubMed]
- Radha, G.; Naik, P.K.; Lopus, M. In vitro characterization and molecular dynamic simulation of shikonin as a tubulin-targeted anticancer agent. Comput. Biol. Med. 2022, 147, 105789. [Google Scholar] [CrossRef] [PubMed]
- Talimarada, D.; Sharma, A.; Holla, H. Identification of dual binding mode of Orthodiffenes towards human topoisomerase-I and alpha-tubulin: Exploring the potential role in anti-cancer activity via in silico study. J. Biomol. Struct. Dyn. 2022, 1–15. [Google Scholar] [CrossRef]
- Majumdar, S.; Basu, D.; Ghosh Dastidar, S. Conformational States of E7010 Is Complemented by Microclusters of Water Inside the α,β-Tubulin Core. J. Chem. Inf. Model. 2019, 59, 2274–2286. [Google Scholar] [CrossRef]
- Pragyandipta, P.; Meher, R.K.; Reddy, P.K.; Pedaparti, R.; Kantevari, S.; Naik, P.K. Structure Based Design of Tubulin Binding 9-Arylimino Noscapinoids: Chemical Synthesis and Experimental Validation Against Breast Cancer Cell Lines. Anal. Chem. Lett. 2022, 12, 29–43. [Google Scholar] [CrossRef]
- Yang, M.-H.; Mao, J.; Zhu, J.-H.; Zhang, H.; Ding, L. Wangzaozin A, a potent novel microtubule stabilizer, targets both the taxane and laulimalide sites on β-tubulin through molecular dynamics simulations. Life Sci. 2022, 301, 120583. [Google Scholar] [CrossRef]

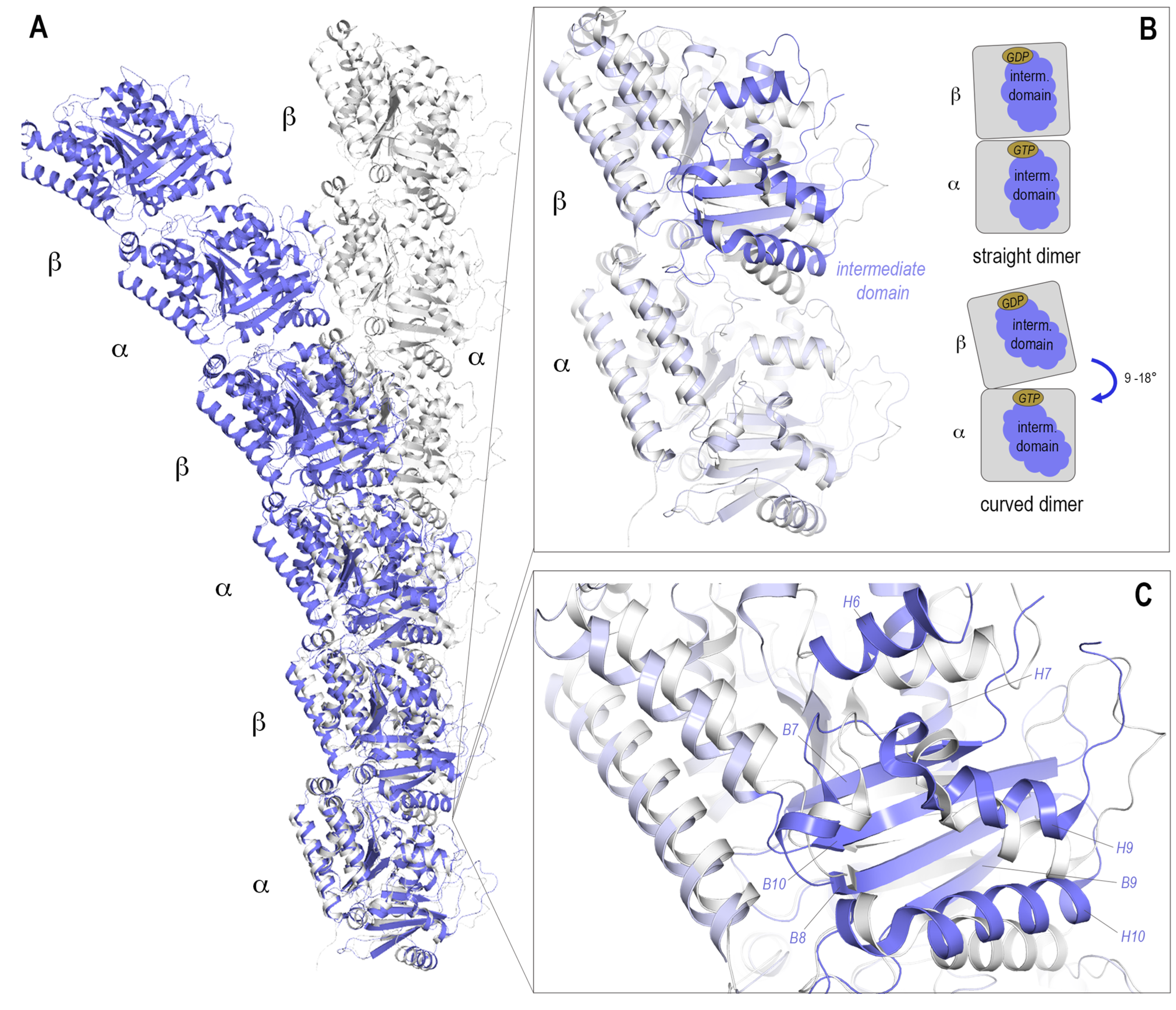
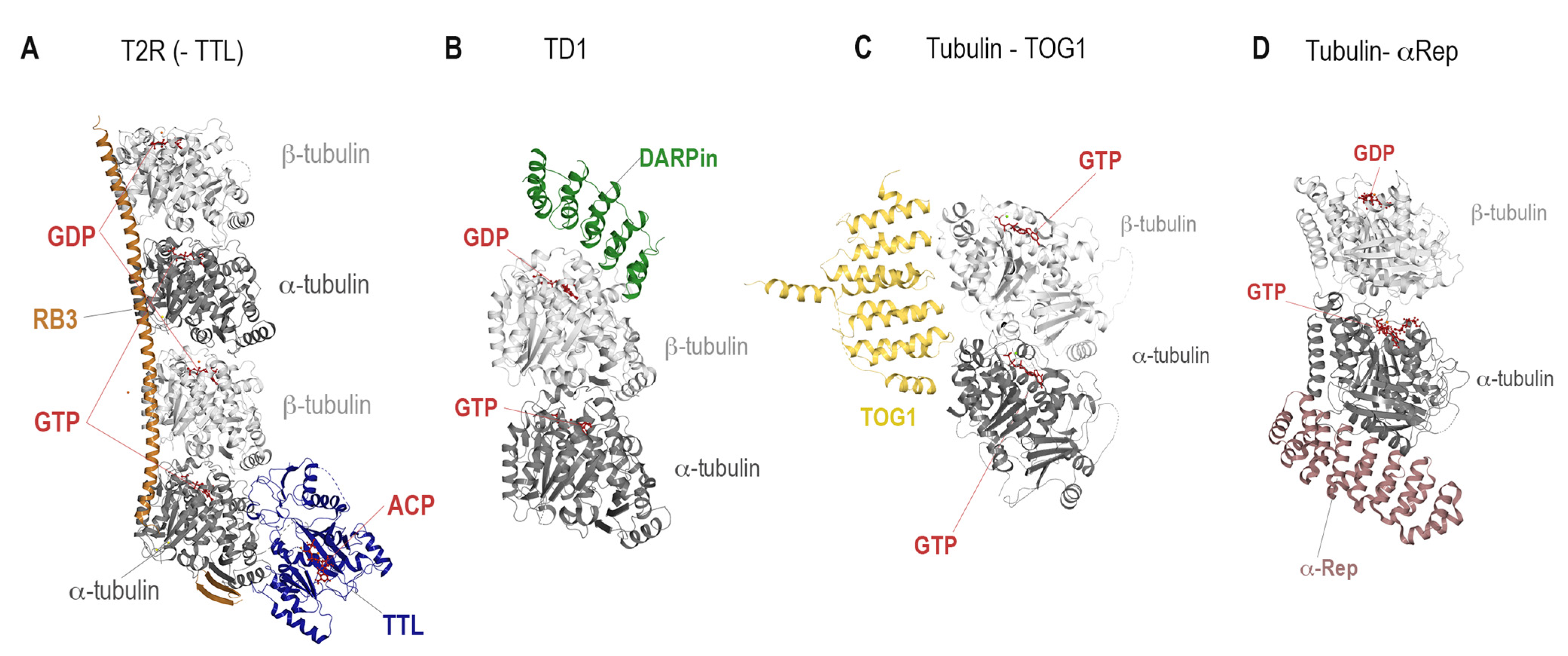

| Binding Site | PDB ID | Resolution (Å) | Crystallization System | Bound Ligand |
|---|---|---|---|---|
| Apo | 5NQU [68] | 1.8 | TD1 | - |
| 3RYC [69] | 2.1 | T2R | - | |
| 4I55 [44] | 2.2 | T2R-TTL | - | |
| Taxane site | 4I4T [44] | 1.8 | T2R-TTL | Zampanolide |
| 5LXT [70] | 1.9 | T2R-TTL | Discodermolide | |
| 6SES [71] | 2.0 | T2R-TTL | B2 | |
| Laulimalide/Peloruside | 4O4H [56] | 2.1 | T2R-TTL | Laulimalide |
| 4O4J [56] | 2.2 | T2R-TTL | Peroluside A | |
| Maytansine | 4TV9 [61] | 2.0 | T2R-TTL | PM060184 |
| 6FJM [72] | 2.1 | T2R-TTL | Disorazole Z | |
| 4TV8 [61] | 2.1 | T2R-TTL | Maytansine | |
| Colchicine | 6S8K [46] | 1.5 | TD1 | Plinabulin |
| 6ZWB [73] | 1.7 | TD1 | Z-SBTub3 photoswitch | |
| 7Z2P [74] | 2.0 | T2R-TTL | Nocodazole | |
| 5M7E [75] | 2.0 | T2R-TTL | BKM120 | |
| 6TH4 [76] | 2.1 | T2R | exo-methylene-nor-colchicine | |
| Vinca | 5IYZ [77] | 1.8 | T2R-TTL | Monomethylauristatin E |
| 5J2T [77] | 2.2 | T2R-TTL | Vinblastine | |
| 5JH7 [67] | 2.3 | T2R-TTL | Eribulin | |
| Pironetin | 5LA6 [62] | 2.1 | T2R-TTL | Pironetin |
| 5FNV [63] | 2.6 | T2R-TTL | Pironetin | |
| Todalam | 5SB3 [12] | 2.2 | T2R-TTL | Todalam precursor 4 |
| 5SB6 [12] | 2.3 | T2R-TTL | Todalam derivative 10 | |
| Gatorbulin | 7ALR [64] | 1.9 | TD1 | Gatorbulin |
| MT Structure | PDBID | Resolution (Å) |
|---|---|---|
| Taxol-stablized MTs | 6WVR [78] | 2.9 |
| Peloruside stabilized MTs | 5SYC [55] | 3.5 |
| Taxol/Peloruside MTs | 5SYE [55] | 3.5 |
| Taxol MTs | 5SYF [55] | 3.5 |
| Zampanolide MTs | 5SYG [55] | 3.5 |
| Undecorated MTs recombinant tubulin | 7SJ7 [79] | 3.8 |
| MD Analysis Metrics | Definition | Examples of Application |
|---|---|---|
| RMSD | The root mean square deviation (RMSD) is a standard measure of the structural distance between coordinates: it measures the average distance between a group of atoms. RMSD values help to evaluate the global structural stability of the system studied in the simulation. | Dash 2022 [119], El-Mernissi 2022 [112], Zhang 2022 [115], Zhao 2022 [120], Radha 2022 [123] |
| RMSF | The root mean square fluctuation (RMSF) represents the quadratic deviation of the atoms in temporal averages. RMSF values help to evaluate the internal structural flexibility of the studied system in the simulation. | Dash 2022 [119], El-Mernissi 2022, Zhang 2022 [115] Radha 2022 [123], Talimarada 2022 [124] |
| Rg | The radius of gyration (Rg) is defined as the mass-weighted root mean square atomic distance from the center-of-mass and can be applied to measure the level of structural compactness of a protein at different time points during the trajectory. | Hadizadeh 2022 [111], El-Mernissi 2022 [112], Zhang 2022 [115], Radha 2022 [123]. Rai 2022 [107] |
| SASA | The solvent accessible surface area (SASA) permits assessment of the overall changes in the tertiary structure of a molecule and its solvent accessibility over the course of the simulation. | El-Mernissi 2022 [112] Rai 2022 [107] |
| 2D interaction analysis | 2D interactions established between the protein and the ligand along the course of the simulations help to identify the residues within the binding site that play an important role in the binding of the ligand to the receptor and to list the ‘hot spots’ between the ligand and the protein. | Basu 2022 [121], Mao 2022 [90], Zhao 2022 [120], Rai 2022 [107], Zhang 2022 [103], Majumdar 2022 [125], Mao 2022 [90], Hadizadeh 2022 [111], Zhang 2022 [115] |
| DSSP | The Define Secondary Structure of Proteins (DSSP) algorithm is the standard method for assigning a secondary structure to amino acids of a protein given the atomic resolution coordinates of the protein. | Mao 2022 [90], Basu 2022 [121] |
| Clustering | Clustering is a data mining technique that allows molecular configurations to be grouped into subsets based on the similarity of their conformations. | Zhang 2022 [115] |
| Binding free energy | The Gibbs free energy (G) provides valuable information about the structure and stability of biomolecules. It is possible to calculate the predicted binding energy (ΔGbind) of a given tubulin-ligand complex using the MD simulation trajectory of this biomolecular association. | Zhao 2022 [120], Zhang 2022 [115], Elhemely 2022 [116], Rai 2022 [107], Radha 2022 [123], Majumdar 2019 [125] |
| PRED | The Per Residue Energy Decomposition (PRED) is a computational tool that is used to obtain the residue-wise contribution to the total binding free energy. It provides information on the key residues that contribute to protein-ligand association, the so-called ‘hot spots’. | Dash 2022 [119], Mao 2022 [90], Zhao 2022 [120], Zhang 2022 [120] |
| CAS | Computational Alanine Scanning (CAS) is a technique that consists of the mutation of amino acids present on the interaction surface between the protein and the ligand to alanine, and the measurement of the difference in binding free energy between the ligand and the native protein and the ligand and the multiple mutated proteins to identify ‘hot spots’. | Neto 2022 [118] |
| PCA | Principal Component Analysis (PCA) is a linear dimensionality reduction tool used in the MD field to map the coordinates of each frame of the trajectory to a linear combination of orthogonal vectors and to investigate the internal modes of motion of the system under study. | Basu 2022 [121] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Peña, H.; Abel, A.-C.; Shevelev, M.; Prota, A.E.; Pieraccini, S.; Horvath, D. Computational Approaches to the Rational Design of Tubulin-Targeting Agents. Biomolecules 2023, 13, 285. https://doi.org/10.3390/biom13020285
Pérez-Peña H, Abel A-C, Shevelev M, Prota AE, Pieraccini S, Horvath D. Computational Approaches to the Rational Design of Tubulin-Targeting Agents. Biomolecules. 2023; 13(2):285. https://doi.org/10.3390/biom13020285
Chicago/Turabian StylePérez-Peña, Helena, Anne-Catherine Abel, Maxim Shevelev, Andrea E. Prota, Stefano Pieraccini, and Dragos Horvath. 2023. "Computational Approaches to the Rational Design of Tubulin-Targeting Agents" Biomolecules 13, no. 2: 285. https://doi.org/10.3390/biom13020285






