Overview of CircRNAs Roles and Mechanisms in Liver Fibrosis
Abstract
:1. Introduction
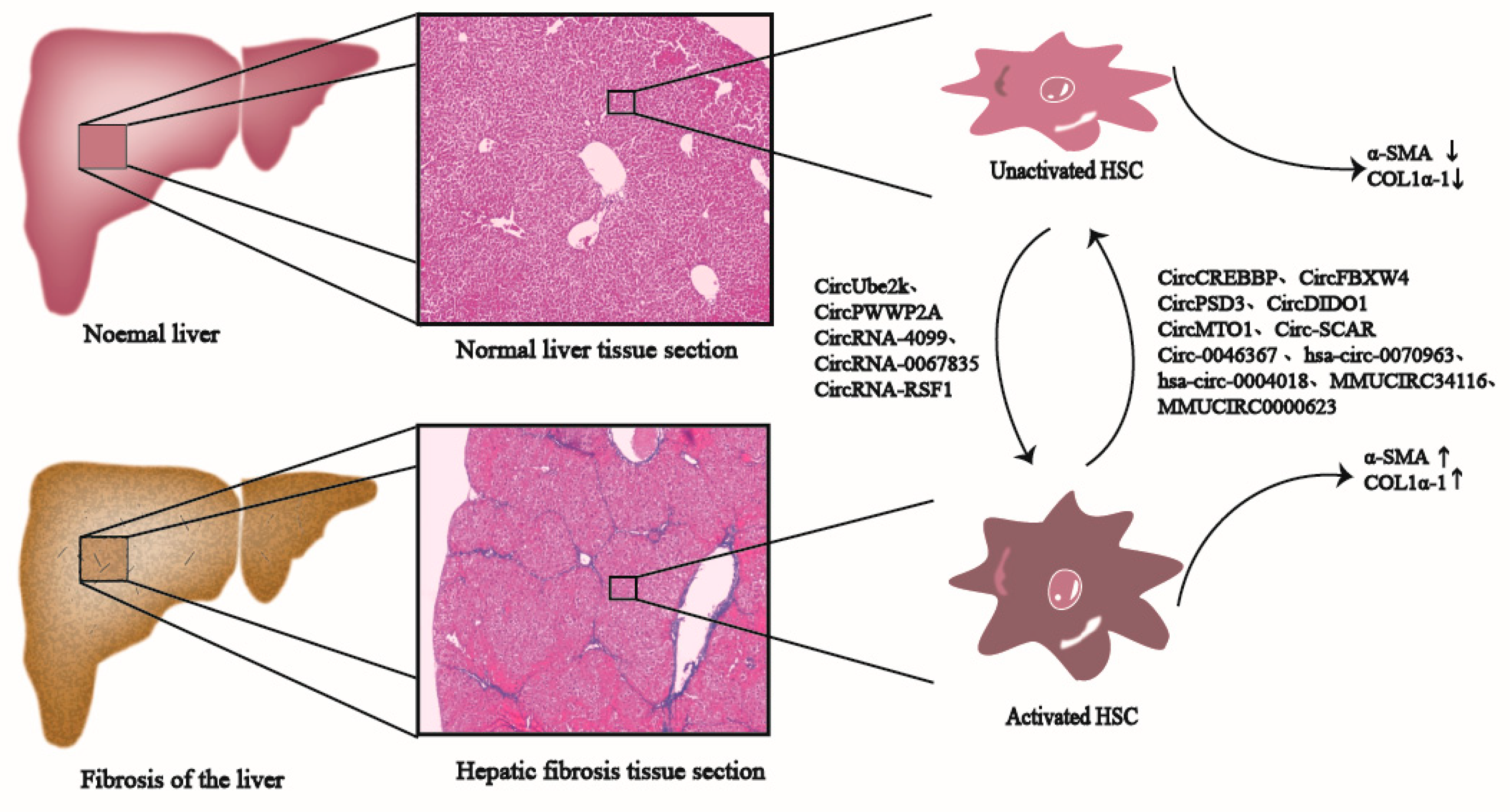
2. Liver Fibrosis
3. CircRNAs Production and Functional Mechanisms
4. Mechanism of CircRNAs in Liver Fibrosis
4.1. CircRNAs Are Involved in Inhibiting Liver Fibrosis
4.1.1. CircRNAs Are Related to Liver Fibrosis through Serving as RNA Scaffolds
CircRNA SCAR
4.1.2. CircRNAs Are Related to Liver Fibrosis by Playing a Role of miRNA-Binding Cernas
CircCREBBP
CircFBXW4
CircPSD3
Hsa_circ_0070963
Hsa_circ_0004018
Mmu_circ_34116
CircDIDO1
CircMTO1
CircRNA-0046367
Mmu_circ_0000623
CircRNA-608
LNCPINT-Derived CircRNAs
MecciRNAs
4.2. CircRNAs Are Involved in Promoting Liver Fibrosis
4.2.1. CircRNAs Are Related to Liver Fibrosis by Playing a Role of miRNAs-Binding CeRNAs
CircUbe2k
CircPWWP2A
CircRNA-4099
CircRNA-0067835
CircRSF1 and Hsa_circ_0071410
CircTUBD1
Hsa_circ_0072765
CircMcph1
CircRNA-0008494
5. Potential Value of CircRNAs in the Treatment of Liver Fibrosis
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, L.; Ma, W.; Wu, L.; Xu, M.; Yang, Y.; Zhang, W.; Sha, W.; Li, H.; Xu, J.; Feng, R.; et al. Glial cell line-derived neurotrophic factor (GDNF) mediates hepatic stellate cell activation via ALK5/Smad signalling. Gut 2019, 68, 2214–2227. [Google Scholar] [CrossRef] [Green Version]
- Breitkopf-Heinlein, K.; Meyer, C.; Konig, C.; Gaitantzi, H.; Addante, A.; Thomas, M.; Wiercinska, E.; Cai, C.; Li, Q.; Wan, F.; et al. BMP-9 interferes with liver regeneration and promotes liver fibrosis. Gut 2017, 66, 939–954. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.Y.; Yuan, W.G.; He, P.; Lei, J.H.; Wang, C.X. Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets. World J. Gastroenterol. 2016, 22, 10512–10522. [Google Scholar] [CrossRef]
- Liu, W.; Feng, R.; Li, X.; Li, D.; Zhai, W. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging 2019, 11, 9569–9580. [Google Scholar] [CrossRef]
- Campana, L.; Iredale, J.P. Regression of Liver Fibrosis. Semin. Liver Dis. 2017, 37, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dong, R.L.; Guo, Y.; He, J.A.; Shao, C.P.; Yi, P.; Yu, F.J.; Gu, D.Y.; Zheng, J.J. CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J. Cell. Mol. Med. 2019, 23, 5486–5496. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, N.; Kawada, N. Role of the Gut-Liver Axis in Liver Inflammation, Fibrosis, and Cancer: A Special Focus on the Gut Microbiota Relationship. Hepatol. Commun. 2019, 3, 456–470. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Sousa, M.A.; Gomez-Moreno, A.Z.; Pineda-Tenor, D.; Sanchez-Ruano, J.J.; Artaza-Varasa, T.; Martin-Vicente, M.; Fernandez-Rodriguez, A.; Martinez, I.; Resino, S. Impact of DARC rs12075 Variants on Liver Fibrosis Progression in Patients with Chronic Hepatitis C: A Retrospective Study. Biomolecules 2019, 9, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, D.A. Reversibility of liver fibrosis. Gastroenterol. Hepatol. 2013, 9, 737–739. [Google Scholar]
- Cobb, B.R.; Valsamakis, A. Chronic Hepatitis B, C, and D. Microbiol. Spectr. 2016, 4, 533. [Google Scholar] [CrossRef]
- Zou, G.L.; Zuo, S.; Lu, S.; Hu, R.H.; Lu, Y.Y.; Yang, J.; Deng, K.S.; Wu, Y.T.; Mu, M.; Zhu, J.J.; et al. Bone morphogenetic protein-7 represses hepatic stellate cell activation and liver fibrosis via regulation of TGF-beta/Smad signaling pathway. World J. Gastroenterol. 2019, 25, 4222–4234. [Google Scholar] [CrossRef] [PubMed]
- Bu, F.T.; Zhu, Y.; Chen, X.; Wang, A.; Zhang, Y.F.; You, H.M.; Yang, Y.; Yang, Y.R.; Huang, C.; Li, J. Circular RNA circPSD3 alleviates hepatic fibrogenesis by regulating the miR-92b-3p/Smad7 axis. Mol. Ther. Nucleic Acids 2021, 23, 847–862. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, E.S.H.; Chen, H.Y.; Man, K.; Go, M.Y.Y.; Huang, X.R.; Lan, H.Y.; Sung, J.J.Y.; Yu, J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 2015, 6, 7325–7338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagan, P.; Sultan, M.; Tachlytski, I.; Safran, M.; Ben-Ari, Z. Both MAPK and STAT3 signal transduction pathways are necessary for IL-6-dependent hepatic stellate cells activation. PLoS ONE 2017, 12, e0176173. [Google Scholar] [CrossRef] [Green Version]
- Xiang, D.M.; Sun, W.; Ning, B.F.; Zhou, T.F.; Li, X.F.; Zhong, W.; Cheng, Z.; Xia, M.Y.; Wang, X.; Deng, X.; et al. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut 2018, 67, 1704–1715. [Google Scholar] [CrossRef]
- Wree, A.; McGeough, M.D.; Inzaugarat, M.E.; Eguchi, A.; Schuster, S.; Johnson, C.D.; Pena, C.A.; Geisler, L.J.; Papouchado, B.G.; Hoffman, H.M.; et al. NLRP3 inflammasome driven liver injury and fibrosis: Roles of IL-17 and TNF in mice. Hepatology 2018, 67, 736–749. [Google Scholar] [CrossRef] [Green Version]
- Molina, M.F.; Abdelnabi, M.N.; Fabre, T.; Shoukry, N.H. Type 3 cytokines in liver fibrosis and liver cancer. Cytokine 2019, 124, 154497. [Google Scholar] [CrossRef]
- Fabre, T.; Molina, M.F.; Soucy, G.; Goulet, J.P.; Willems, B.; Villeneuve, J.P.; Bilodeau, M.; Shoukry, N.H. Type 3 cytokines IL-17A and IL-22 drive TGF-β-dependent liver fibrosis. Sci. Immunol. 2018, 3, eaar7754. [Google Scholar] [CrossRef] [Green Version]
- Friedman, S.L. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008, 88, 125–172. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.D.; Bu, F.T.; Li, X.F.; Chen, Y.; Zhu, S.; Wang, J.N.; Chen, S.Y.; Sun, Y.Y.; Pan, X.Y.; et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 2020, 10, 4851–4870. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Song, F.M.; Lei, X.; Li, J.T.; Li, F.; Tan, H.B. hsa_circ_0004018 suppresses the progression of liver fibrosis through regulating the hsa-miR-660-3p/TEP1 axis. Aging 2020, 12, 11517–11529. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xu, Q.G.; Wang, Z.G.; Yang, Y.; Zhang, L.; Ma, J.Z.; Sun, S.H.; Yang, F.; Zhou, W.P. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018, 68, 1214–1227. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Hentze, M.W.; Preiss, T. Circular RNAs: Splicing’s enigma variations. EMBO J. 2013, 32, 923–925. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Tsukahara, T. A View of Pre-mRNA Splicing from RNase R Resistant RNAs. Int. J. Mol. Sci. 2014, 15, 9331–9342. [Google Scholar] [CrossRef] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.Z.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Lener, T.; Gimona, M.; Aigner, L.; Börger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; Del Portillo, H.A.; et al. Applying extracellular vesicles based therapeutics in clinical trials—An ISEV position paper. J. Extracell. Vesicles 2015, 4, 30087. [Google Scholar] [CrossRef] [Green Version]
- Yao, T.; Chen, Q.Q.; Fu, L.Y.; Guo, J.M. Circular RNAs: Biogenesis, properties, roles, and their relationships with liver diseases. Hepatol. Res. 2017, 47, 497–504. [Google Scholar] [CrossRef]
- Yang, L.; Wilusz, J.E.; Chen, L.L. Biogenesis and Regulatory Roles of Circular RNAs. Annu. Rev. Cell Dev. Biol. 2022, 38, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Reckman, Y.J.; Aufiero, S.; van den Hoogenhof, M.M.; van der Made, I.; Beqqali, A.; Koolbergen, D.R.; Rasmussen, T.B.; van der Velden, J.; Creemers, E.E.; et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ. Res. 2016, 119, 996–1003. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Dai, Z.H.; Liu, F.C.; Guo, X.G.; Ge, C.M.; Ding, J.; Liu, H.; Yang, F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 2019, 45, 155–167. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015, 29, 2168–2182. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.Q.; An, O.; Ren, X.; Song, Y.Y.; Tang, Z.J.; Ke, X.Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Molias, F.B.; et al. ADARs act as potent regulators of circular transcriptome in cancer. Nat. Commun. 2022, 13, 1508. [Google Scholar] [CrossRef]
- Kokot, K.E.; Kneuer, J.M.; John, D.; Rebs, S.; Mobius-Winkler, M.N.; Erbe, S.; Muller, M.; Andritschke, M.; Gaul, S.; Sheikh, B.N.; et al. Reduction of A-to-I RNA editing in the failing human heart regulates formation of circular RNAs. Basic Res. Cardiol. 2022, 117, 32. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.Y.; Zhang, J.L.; Yang, L.; Chen, L.L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef] [Green Version]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; Robinson, D.R.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019, 176, 831–843.e822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Xu, J.; Ji, L.; Liang, Y.; Wan, Z.; Zheng, W.; Song, X.; Gorshkov, K.; Sun, Q.; Lin, H.; Zheng, X.; et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target. Ther. 2020, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.Y.; Liu, J.Y.; Deng, H.; Ma, R.Y.; Liao, J.Y.; Liang, H.X.; Hu, J.X.; Li, J.Q.; Guo, Z.Y.; Cai, J.C.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93.e22. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, R.; Li, J.; Tang, S.; Li, S.; Tong, Q. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int. J. Nanomed. 2021, 16, 2803–2818. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.N.; Liu, E.; Yang, Z.G.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.Y.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.L.; Zhong, G.L.; Yu, B.; Hu, W.C.; Dai, L.M.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Yang, Y.B.; Gao, X.Y.; Zhang, M.L.; Yan, S.; Sun, C.J.; Xiao, F.Z.; Huang, N.N.; Yang, X.S.; Zhao, K.; Zhou, H.K.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018, 110, 304–315. [Google Scholar] [CrossRef] [Green Version]
- Zuo, L.; Zhang, L.; Zu, J.; Wang, Z.; Han, B.; Chen, B.L.; Cheng, M.J.; Ju, M.Z.; Li, M.Y.; Shu, G.F.; et al. Circulating Circular RNAs as Biomarkers for the Diagnosis and Prediction of Outcomes in Acute Ischemic Stroke. Stroke 2020, 51, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Li, S.N.; Hu, W.D.; Deng, F.; Chen, S.F.; Zhu, P.Y.; Wang, M.X.; Chen, X.L.; Wang, Y.; Hu, X.J.; Zhao, B.; et al. Identification of Circular RNA hsa_circ_0001599 As a Novel Biomarker for Large-Artery Atherosclerotic Stroke. DNA Cell Biol. 2021, 40, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zheng, Y.L.; Hao, D.Z.; Jin, X.S.; Luo, Q.Z.; Guo, Y.T.; Li, D.X.; Xi, W.; Xu, Y.; Chen, Y.S.; et al. Blood circRNAs as biomarkers for the diagnosis of community-acquired pneumonia. J. Cell. Biochem. 2019, 120, 16483–16494. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.R.; Hu, S.; Bu, F.T.; Li, H.; Huang, C.; Meng, X.M.; Zhang, L.; Lv, X.W.; Li, J. Circular RNA CREBBP Suppresses Hepatic Fibrosis Via Targeting the hsa-miR-1291/LEFTY2 Axis. Front. Pharmacol. 2021, 12, 2515. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Chen, X.; Wang, J.N.; Xu, J.J.; Wang, A.; Li, J.J.; Wu, S.; Wu, Y.Y.; Li, X.F.; Huang, C.; et al. Circular RNA circUbe2k promotes hepatic fibrosis via sponging miR-149-5p/TGF-β2 axis. FASEB J. 2021, 35, e21622. [Google Scholar] [CrossRef]
- Zhou, Y.P.; Lv, X.Y.; Qu, H.; Zhao, K.K.; Fu, L.Y.; Zhu, L.W.; Ye, G.L.; Guo, J.M. Differential expression of circular RNAs in hepatic tissue in a model of liver fibrosis and functional analysis of their target genes. Hepatol. Res. 2019, 49, 324–334. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yuan, B.Y.; Wu, Z.F.; Dong, Y.Y.; Zhang, L.; Zeng, Z.C. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene 2017, 629, 35–42. [Google Scholar] [CrossRef]
- Ji, D.; Chen, G.F.; Wang, J.C.; Ji, S.H.; Wu, X.W.; Lu, X.J.; Chen, J.L.; Li, J.T. Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging 2020, 12, 1643–1655. [Google Scholar] [CrossRef]
- Zhou, Y.P.; Lv, X.Y.; Qu, H.; Zhao, K.K.; Fu, L.Y.; Zhu, L.W.; Ye, G.L.; Guo, J.M. Preliminary screening and functional analysis of circular RNAs associated with hepatic stellate cell activation. Gene 2018, 677, 317–323. [Google Scholar] [CrossRef]
- Ma, L.; Wei, J.F.; Zeng, Y.L.; Liu, J.P.; Xiao, E.H.; Kang, Y.H.; Kang, Y. Mesenchymal stem cell-originated exosomal circDIDO1 suppresses hepatic stellate cell activation by miR-141-3p/PTEN/AKT pathway in human liver fibrosis. Drug Deliv. 2022, 29, 440–453. [Google Scholar] [CrossRef]
- Jin, H.; Li, C.X.; Dong, P.H.; Huang, J.T.; Yu, J.L.; Zheng, J.J. Circular RNA cMTO1 Promotes PTEN Expression Through Sponging miR-181b-5p in Liver Fibrosis. Front. Cell Dev. Biol. 2020, 8, 714. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Liu, X.; Li, W.; Wang, L. Exosomes derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis via activating autophagy. Hum. Exp. Toxicol. 2020, 39, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Chen, J.N.; Sun, F.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxida. Med. Cell. Longev. 2017, 2017, 3960197. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.X.; Li, J.Z.; Li, Q.; Xu, M.Y.; Li, H.Y. CircRNA608-microRNA222-PINK1 axis regulates the mitophagy of hepatic stellate cells in NASH related fibrosis. Biochem. Biophys. Res. Commun. 2022, 610, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Cao, Y.; Guo, C.J.; Guo, X.Y.; He, Y.F.; Xu, Q.Y.; Shen, F.; Pan, Q. Profile analysis and functional modeling identify circular RNAs in nonalcoholic fatty liver disease as regulators of hepatic lipid metabolism. Front. Genet. 2022, 13, 884037. [Google Scholar] [CrossRef]
- Liu, B.Q.; Tian, Y.S.; He, J.; Gu, Q.X.; Jin, B.H.; Shen, H.; Li, W.Q.; Shi, L.; Yu, H.; Shan, G.; et al. The potential of mecciRNA in hepatic stellate cell to regulate progression of nonalcoholic hepatitis. J. Transl. Med. 2022, 20, 393. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.L.; Li, J.X.; Hu, S.S.; Deng, Y.Q.; Yin, H.; Bao, X.C.; Zhang, Q.C.; Wang, G.; Wang, B.L.; et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China-Life Sci. 2020, 63, 1429–1449. [Google Scholar] [CrossRef]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Li, Y.L.; Gao, X.J.; Wang, Z.H.; Liu, W.; Xu, F.; Hu, Y.J.; Li, Y.N.; Shi, L. Circular RNA 4099 aggravates hydrogen peroxide-induced injury by down-regulating microRNA-706 in L02 cells (Retracted article. See vol. 308, 2022). Life Sci. 2020, 241, 116826. [Google Scholar] [CrossRef]
- Zhu, L.; Ren, T.; Zhu, Z.; Cheng, M.; Mou, Q.; Mu, M.; Liu, Y.; Yao, Y.; Cheng, Y.; Zhang, B.; et al. Thymosin-β4 Mediates Hepatic Stellate Cell Activation by Interfering with CircRNA-0067835/miR-155/FoxO3 Signaling Pathway. Cell Physiol. Biochem. 2018, 51, 1389–1398. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yuan, B.Y.; Chen, G.W.; Zhang, L.; Zhuang, Y.; Niu, H.; Zeng, Z.C. Circular RNA RSF1 promotes inflammatory and fibrotic phenotypes of irradiated hepatic stellate cell by modulating miR-146a-5p. J. Cell. Physiol. 2020, 235, 8270–8282. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Zhang, L.; Wang, B.A.; Zhang, G.C.; Liu, J.; Wu, Z.F.; Du, S.S.; Zeng, Z.C. CircTUBD1 Regulates Radiation-induced Liver Fibrosis Response via a circTUBD1/micro-203a-3p/Smad3 Positive Feedback Loop. J. Clin. Transl. Hepatol. 2022, 10, 680–691. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Guo, X.; Zhang, R.; Yan, C. Hsa_circ_0072765 knockdown inhibits proliferation, activation and migration in transforming growth factor-beta (TGF-β)-induced hepatic stellate cells (HSCs) by the miR-197-3p/TRPV3 axis. Histol. Histopathol. 2023, 17, 18–586. [Google Scholar]
- Xu, J.J.; Chen, X.; Zhu, S.; Jiang, L.F.; Ma, W.X.; Chen, S.Y.; Meng, X.M.; Huang, C.; Li, J. Myc-mediated circular RNA circMcph1/miR-370-3p/Irak2 axis is a progressive regulator in hepatic fibrosis. Life Sci. 2023, 312, 24. [Google Scholar] [CrossRef]
- Li, B.B.; Zhou, J.M.; Luo, Y.Y.; Tao, K.G.; Zhang, L.F.; Zhao, Y.; Lin, Y.; Zeng, X.; Yu, H.Y. Suppressing circ_0008494 inhibits HSCs activation by regulating the miR-185-3p/Col1a1 axis. Front. Pharmacol. 2022, 13, 3002. [Google Scholar] [CrossRef] [PubMed]
- Petrij, F.; Giles, R.H.; Dauwerse, H.G.; Saris, J.J.; Hennekam, R.C.; Masuno, M.; Tommerup, N.; van Ommen, G.J.; Goodman, R.H.; Peters, D.J.; et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 1995, 376, 348–351. [Google Scholar] [CrossRef]
- Jia, D.S.; Augert, A.; Kim, D.W.; Eastwood, E.; Wu, N.; Ibrahim, A.H.; Kim, K.B.; Dunn, C.T.; Pillai, S.P.S.; Gazdar, A.F.; et al. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov. 2018, 8, 1422–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menke, L.A.; Gardeitchik, T.; Hammond, P.; Heimdal, K.R.; Houge, G.; Hufnagel, S.B.; Ji, J.; Johansson, S.; Kant, S.G.; Kinning, E.; et al. Further delineation of an entity caused by CREBBP and EP300 mutations but not resembling Rubinstein-Taybi syndrome. Am. J. Med. Genet. A 2018, 176, 862–876. [Google Scholar] [CrossRef]
- Sadeghi, H.; Esmkhani, S.; Pirjani, R.; Amin-Beidokhti, M.; Gholami, M.; Azizi Tabesh, G.; Ghasemi, M.R.; Gachkar, L.; Mirfakhraie, R. CREB-binding protein (CREBBP) and preeclampsia: A new promising target gene. Mol. Biol. Rep. 2021, 48, 2117–2122. [Google Scholar] [CrossRef]
- Hellemans, J.; Preobrazhenska, O.; Willaert, A.; Debeer, P.; Verdonk, P.C.; Costa, T.; Janssens, K.; Menten, B.; Van Roy, N.; Vermeulen, S.J.; et al. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 2004, 36, 1213–1218. [Google Scholar] [CrossRef]
- Harrington, L.; McPhail, T.; Mar, V.; Zhou, W.; Oulton, R.; Bass, M.B.; Arruda, I.; Robinson, M.O. A mammalian telomerase-associated protein. Science 1997, 275, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Barnard, A.; Moch, A.; Saab, S. Relationship between Telomere Maintenance and Liver Disease. Gut Liver 2019, 13, 11–15. [Google Scholar] [CrossRef] [Green Version]
- Ji, D.; Li, B.; Shao, Q.; Li, F.; Li, Z.B.; Chen, G.F. MiR-22 Suppresses BMP7 in the Development of Cirrhosis. Cell. Physiol. Biochem. 2015, 36, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Li, J.X.; Wang, H.M.; Su, X.P.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.Y.; et al. Circular RNA circMTO1 Acts as the Sponge of MicroRNA-9 to Suppress Hepatocellular Carcinoma Progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.J.; Wu, C.Z.; Xu, Z.Q.; Xia, P.; Dong, P.H.; Chen, B.C.; Yu, F.J. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol. Cell. Biochem. 2015, 398, 1–9. [Google Scholar] [CrossRef]
- Qu, Y.; Zhang, Q.D.; Cai, X.B.; Li, F.; Ma, Z.Z.; Xu, M.Y.; Lu, L.E. Exosomes derived from miR-181-5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J. Cell. Mol. Med. 2017, 21, 2491–2502. [Google Scholar] [CrossRef]
- Lawrence, Y.A.; Dangott, L.J.; Rodrigues-Hoffmann, A.; Steiner, J.M.; Suchodolski, J.S.; Lidbury, J.A. Proteomic analysis of liver tissue from dogs with chronic hepatitis. PLoS ONE 2018, 13, e0208394. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A.L.; Hannappel, E.; Kleinman, H.K. Thymosin beta4: Actin-sequestering protein moonlights to repair injured tissues. Trends Mol. Med. 2005, 11, 421–429. [Google Scholar] [CrossRef]
- Adachi, M.; Osawa, Y.; Uchinami, H.; Kitamura, T.; Accili, D.; Brenner, D.A. The forkhead transcription factor FoxO1 regulates proliferation and transdifferentiation of hepatic stellate cells. Gastroenterology 2007, 132, 1434–1446. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, B.; Levin, W. Late radiation-related fibrosis: Pathogenesis, manifestations, and current management. Semin. Radiat. Oncol. 2003, 13, 274–289. [Google Scholar] [CrossRef]
- Munoz-Schuffenegger, P.; Ng, S.; Dawson, L.A. Radiation-Induced Liver Toxicity. Semin. Radiat. Oncol. 2017, 27, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.B.; Lv, Q.; Wong, C.K.A.; Hu, S.; Fu, C.; Hua, Z.; Cai, G.P.; Li, G.X.; Yang, B.B.; Zhang, Y. The Effect of Central Loops in miRNA:MRE Duplexes on the Efficiency of miRNA-Mediated Gene Regulation. PLoS ONE 2008, 3, e1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, L.H.; Yang, W.J.; Yao, J.Y.; Wang, H.; Zhu, J.; Jin, A.L.; Liu, T.; Wang, B.L.; Zhou, J.; Fan, J.; et al. The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark. Med. 2021, 15, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Guo, J.; Zhao, B.; Zhang, Z.; Zhu, J.; Liu, F. CircCHD2/miR-200b-3p/HLF Axis Promotes Liver Cirrhosis. J. Environ. Pathol. Toxicol. Oncol. 2022, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Deng, T.; Ge, S.H.; Liu, Y.; Bai, M.; Zhu, K.G.; Fan, Q.; Li, J.L.; Ning, T.; Tian, F.; et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019, 38, 2844–2859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.Y.; Liu, W.; Zou, Y.; Wang, G.S.; Deng, Y.N.; Luo, J.Y.; Zhang, Y.C.; Li, H.; Zhang, Q.; Yang, Y.; et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 2019, 40, 432–445. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [Green Version]

| CircRNAs | Targets | Biological Function | Expression Pattern | Ref. |
|---|---|---|---|---|
| Inhibition of liver fibrosis | ||||
| CircRNA SCAR | ATP5B | CircRNA SCAR overexpression clearly inhibit the contractility, collagen and α-SMA expression in NASH fibroblasts, thereby alleviating the fibrotic phenotype. | decrease | [46] |
| CircCREBBP | hsa-miR-1291 | CircCREBBP, as a sponge for hsa-miR-1291, promotes the expression of LEFTY2 and inhibits the activation of hepatic stellate cells. | decrease | [54] |
| CircFBXW4 | miR-18b-3p | CircFBXW4, as a miRNA sponge for miR-18b-3p, to regulate the expression of FBXW7, inhibiting the activation of hepatic stellate cells. | decrease | [21] |
| CircPSD3 | miR-92b-3p | CircPSD3 can inhibit HSC activation and proliferation by targeting the miR-92b-3p/Smad7 axis to alleviate the formation of liver fibrosis. | decrease | [12] |
| Hsa_circ_0070963 | miR-223-3p | Hsa_circ_0070963, as a miR-223-3p sponge, inhibites HSC activation via regulation of miR-223-3p and LEMD3 in liver fibrosis. | decrease | [58] |
| Hsa_circ_0004018 | hsa-miR-660-3p | Hsa_circ_0004018 acts as a sponge of hsa-miR-660-3p, and then targetedly suppress the expression of TEP1 to inhibit the activation of HSCs. | decrease | [22] |
| Mmu_circ_34116 | miR-22 | Mmu_circ_34116 inhibits the activation of HSCs and fibrosis by mmu_circ_34116/miR-22/BMP7 signal axis. | decrease | [59] |
| CircDIDO1 | miR-141-3p | Extracellular circDIDO1 restraines HSC activation by miR-141-3p/PTEN/AKT pathway in liver fibrosis. | decrease | [60] |
| CircMTO1(Hsa_circ_0007874) | miR-17-5p, miR-181b-5p | CircMTO1 can inhibit TGF-β1-induced HSC activation by targeting miR-17-5p and Smad7 or miR-181b-5p/PTEN/AKT cascade. | decrease | [6,61] |
| CircRNA-0046367 | miR-34a | CircRNA-0046367 promotes the expression of CPT2 and ACBD3 by promoting PPARα through CircRNA-0046367/miR-34a/PPARα axis, and reduces steatosis of hepatocytes. | decrease | [63] |
| Mmu_circ_0000623 | miR-125 | Mmu_circ_0000623-modified ADSCs, significantly suppressed CCl4-induced liver fibrosis by promoting autophagy activation through interacting with the miR-125/ATG4D. | decrease | [62] |
| CircRNA-608 | miR-222 | CircRNA-608 might promote PINK1-mediated mitophagy to slow down NASH-related liver fibrosis though inhibiting miR-222 in lipotoxic HSCs. | decrease | [64] |
| LNCPINT-derived CircRNAs (Circ_0001452, Circ_0001453, and Circ_0001454) | miR-466i-3p, miR-669c-3p | Loss of LNCPINT-derived CircRNAs may underlie NAFLD via miR-466i-3p and miR-669c-3p-dependent inactivation of the AMPK signaling pathway. | decrease | [65] |
| MecciRNAs (Hsa_circ_0089761 and Hsa_circ_0089763) | miR-642a-5p, miR-1248, miR-670-3p and miR-1224-3p | Hsa_ circ_0089761 and hsa_circ_0089763 could function as competing for endogenous RNAs (ceRNAs) to regulate fibrosis-related signals. | decrease | [66] |
| MecciRNAs (Hsa_circ_0089762 and Hsa_circ_0008882_ | Hsa_circ_0089762 and hsa_circ_0008882 might act as molecular scaffolds to regulate specific complex functions or serve as molecular chaperones in the folding of mitochondria-imported proteins. | decrease | [67,68] | |
| Promotion of liver fibrosis | ||||
| CircUbe2k(Mmu_circ_0001350) | miR-149-5p | CircUbe2k could promote the activation of HSCs and the progress of HF through the CircUbe2k/miR-149-5p/TGF-β2 axis. | increase | [55] |
| CircPWWP2A | miR-203, miR-223 | CircPWWP2A promoted the activation and proliferation of HSCs via sponging miR-203 and miR-223, subsequently enhanced the expression of Fstl1 and Tlr4, respectively, and eventually promoted hepatic fibrosis. | increase | [4] |
| CircRNA-4099 | miR-706 | CircRNA-4099 aggravated H2O2-induced damage and fibrosis by inhibiting miR-706 through triggering keap1/Nrf2 and p38MAPK in L02 cell lines. | increase | [69] |
| CircRNA-0067835 | miR-155 | CircRNA-0067835 can promote liver fibrosis progression by acting asa sponge of miR-155 to promote FOXO3a and AKT expression in LX-2 cells. | increase | [70] |
| CircRSF1 | miR-146a-5p | CircRSF1 can promote the activation of HSC cells by the CircRSF1/miR-146a-5p/RAC1 axis in irradiated LX-2 cells. | increase | [71] |
| Hsa_circ_0071410 | miR-9-5p | Hsa_circ_0071410 inhibits the proliferation of LX-2 by Hsa_circ_0071410/miR-9-5p in irradiated LX-2cells. | increase | [57] |
| CircTUBD1(Hsa_circ_0044897) | miR-203a-3p | CircTUBD1 regulated the activation and fibrosis response of LX-2 cells induced by irradiation via circTUBD1/miR-203a-3p/Smad3 axis. | increase | [72] |
| Hsa_circ_0072765 | miR-197-3p | Hsa_circ_0072765 promotes the progression of HSCs and liver fibrosis induced by TGF-β through decreasing the expression of miR-197-3p and inducing the expression of TRPV3. | increase | [73] |
| CircMcph1 | miR-370-3p | CircMcph1 could regulate the expression of Irak2 by sponging miR-370-3p. | increase | [74] |
| CircRNA-0008494 | miR-185-3p | CircRNA-0008494 can regulate activation, proliferation, migration and apoptosis of HSCs by CircRNA-0008494/miR-185-3p/Col1a1 axis. | increase | [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Tong, J.; Li, Y.; Qiu, X.; Chen, A.; Chang, C.; Yu, G. Overview of CircRNAs Roles and Mechanisms in Liver Fibrosis. Biomolecules 2023, 13, 940. https://doi.org/10.3390/biom13060940
Wang G, Tong J, Li Y, Qiu X, Chen A, Chang C, Yu G. Overview of CircRNAs Roles and Mechanisms in Liver Fibrosis. Biomolecules. 2023; 13(6):940. https://doi.org/10.3390/biom13060940
Chicago/Turabian StyleWang, Gaiping, Jiahui Tong, Yingle Li, Xianglei Qiu, Anqi Chen, Cuifang Chang, and Guoying Yu. 2023. "Overview of CircRNAs Roles and Mechanisms in Liver Fibrosis" Biomolecules 13, no. 6: 940. https://doi.org/10.3390/biom13060940






