Potential Causal Association between Plasma Metabolites, Immunophenotypes, and Female Reproductive Disorders: A Two-Sample Mendelian Randomization Analysis
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Design and Ethical Approval
2.2. Genetic Instruments for Metabolites and Immune Traits
2.3. Instrumental Variables (IVs) Selection
2.4. Statistical Analysis
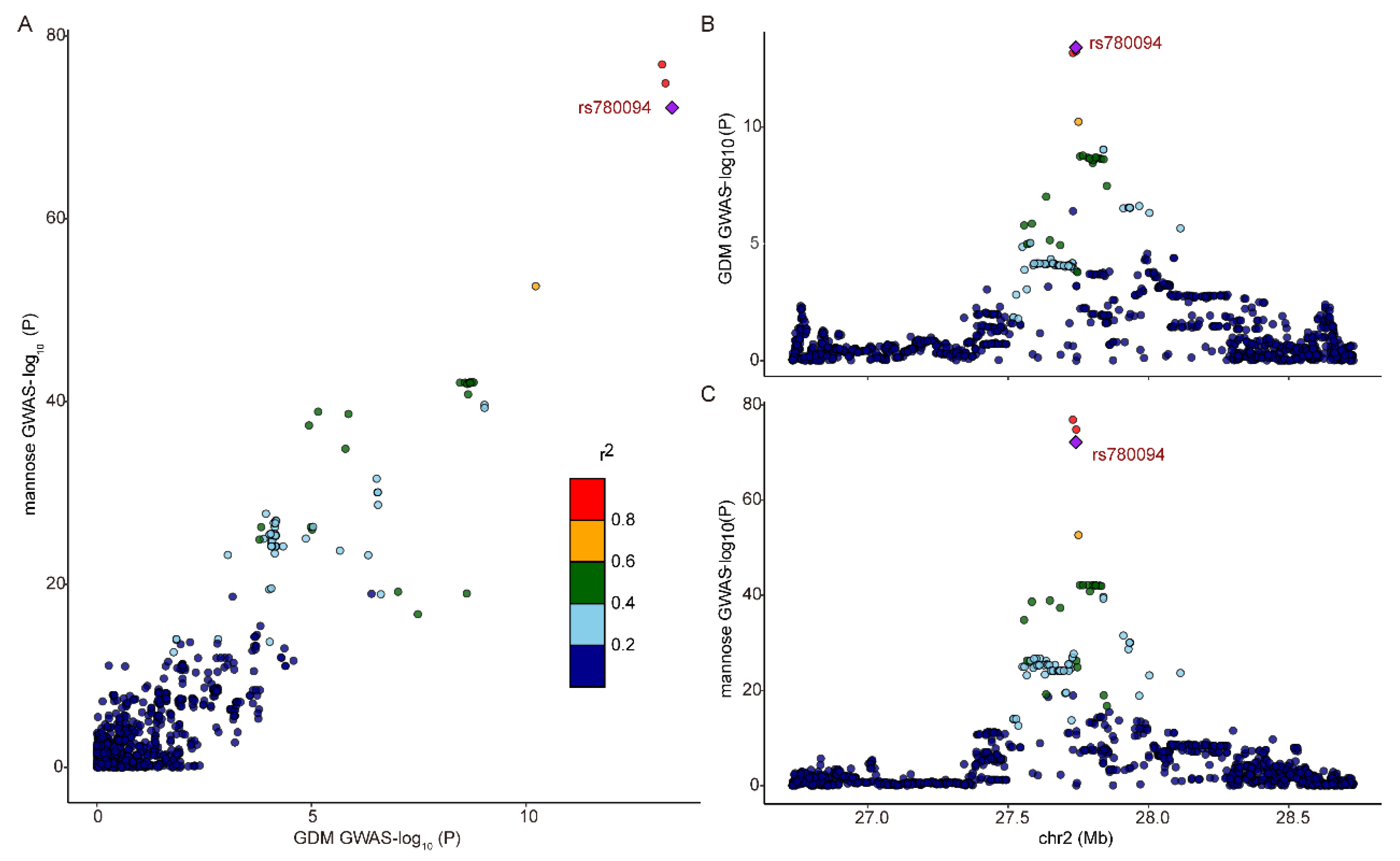
2.5. Colocalization Analyses on Exposure with Outcome
2.6. Metabolic Pathway Analysis
3. Results
3.1. Strength of the Instrumental Variables (IVs)
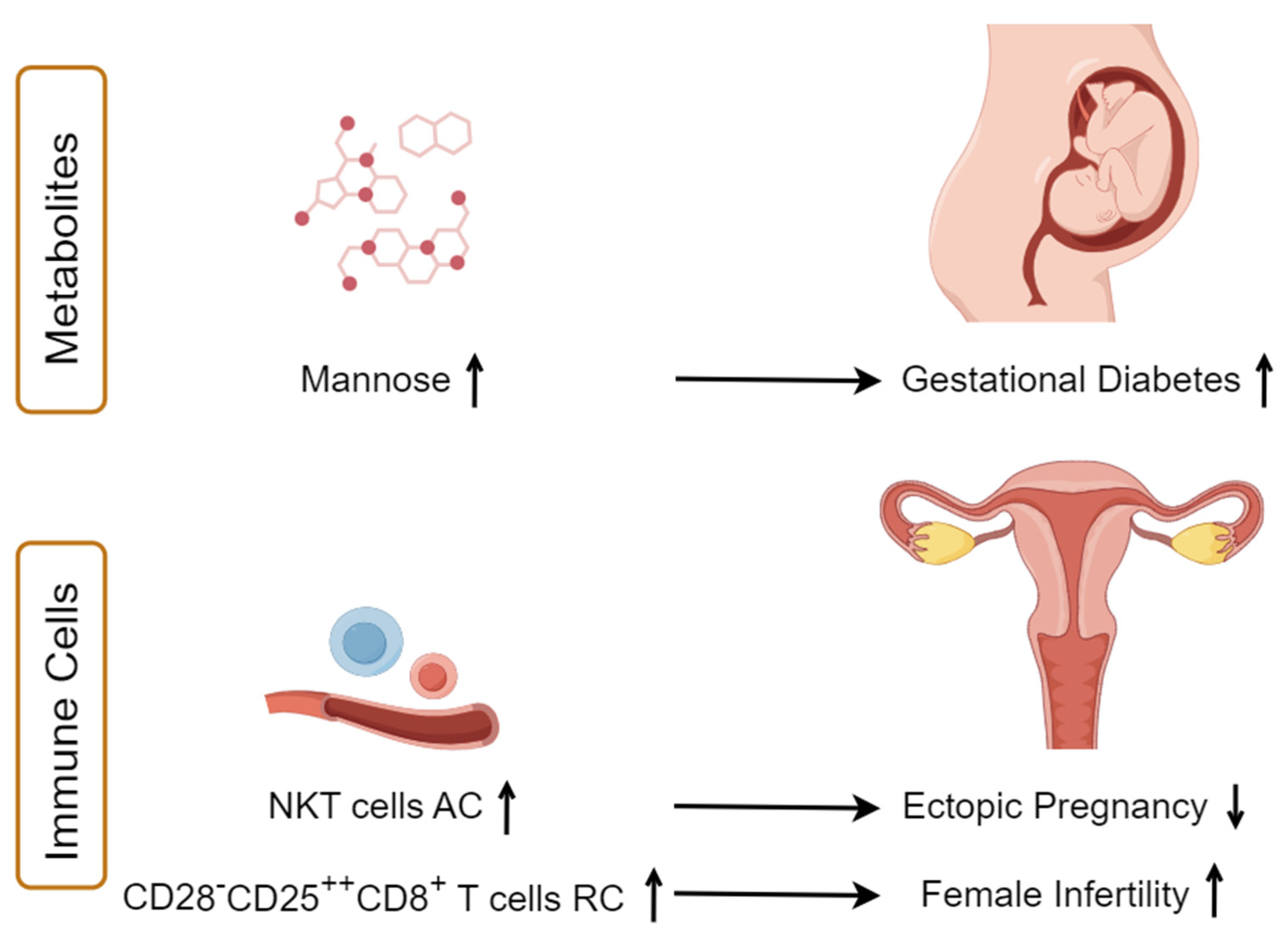
3.2. Identifying the Causal Effect of Immunophenotypes on Female Reproductive Disorders
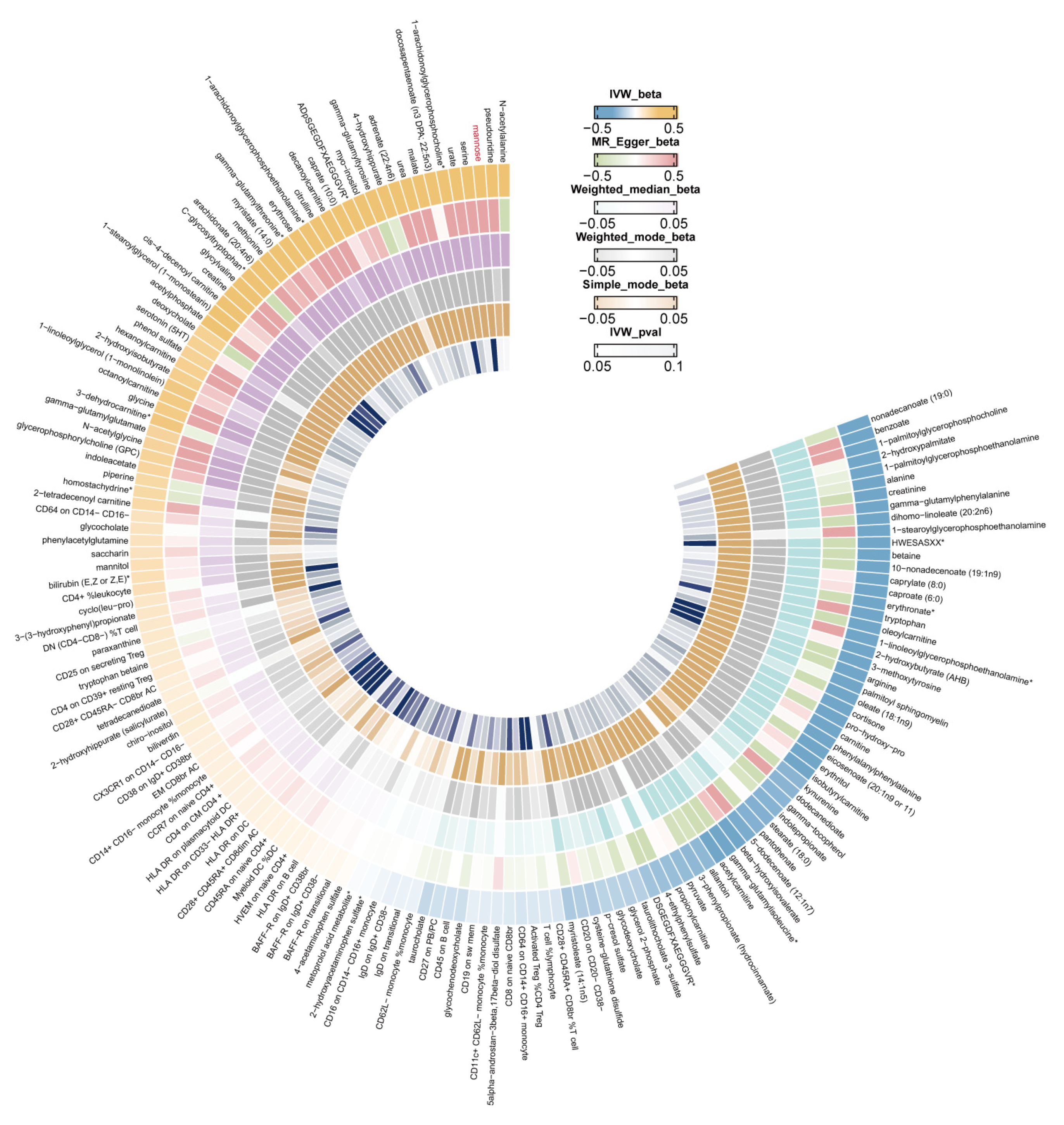
3.3. Identifying the Causal Effect of Metabolites on Female Reproductive Disorders
3.4. Metabolic Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Skakkebaek, N.E.; Jørgensen, N.; Andersson, A.M.; Juul, A.; Main, K.M.; Jensen, T.K.; Toppari, J. Populations, decreasing fertility, and reproductive health. Lancet 2019, 393, 1500–1501. [Google Scholar] [CrossRef] [PubMed]
- Alesi, S.; Ghelani, D.; Rassie, K.; Mousa, A. Metabolomic Biomarkers in Gestational Diabetes Mellitus: A Review of the Evidence. Int. J. Mol. Sci. 2021, 22, 5512. [Google Scholar] [CrossRef]
- Alesi, S.; Ghelani, D.; Mousa, A. Metabolomic Biomarkers in Polycystic Ovary Syndrome: A Review of the Evidence. Semin. Reprod. Med. 2021, 39, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Zhang, T.; Wu, L.; Li, T.C.; Wang, C.C.; Chung, J.P.W. Metabolomic markers of biological fluid in women with reproductive failure: A systematic review of current literatures. Biol. Reprod. 2022, 106, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.-J.; Yang, H.-L.; Mei, J.; Chang, K.-K.; Lu, H.; Lai, Z.-Z.; Shi, J.-W.; Wang, X.-H.; Wu, K.; Zhang, T.; et al. Fructose-1,6-bisphosphate prevents pregnancy loss by inducing decidual COX-2 macrophage differentiation. Sci. Adv. 2022, 8, eabj2488. [Google Scholar] [CrossRef] [PubMed]
- Gojnic-Dugalic, M.; Stefanovic, K.; Stefanovic, A.; Jotic, A.; Lalic, N.; Petronijevic-Vrzic, S.; Petronijevic, M.; Milicic, T.; Lukic, L.; Todorovic, J.; et al. Distribution of normal and pathological OGTTs among pregnant population and non-pregnant women with PCOS—The cross-sectional study. Medicine 2021, 100, e27232. [Google Scholar] [CrossRef]
- Saeedi, M.; Cao, Y.; Fadl, H.; Gustafson, H.; Simmons, D. Increasing prevalence of gestational diabetes mellitus when implementing the IADPSG criteria: A systematic review and meta-analysis. Diabetes Res. Clin. Pract. 2021, 172, 108642. [Google Scholar] [CrossRef]
- Najafi, L.; Abedini, A.; Kadivar, M.; Khajavi, A.; Bordbar, A.; Noohi, A.H.; Mashak, B.; Hashemnejad, M.; Khamseh, M.E.; Malek, M. Gestational diabetes mellitus: The correlation between umbilical coiling index, and intrapartum as well as neonatal outcomes. J. Diabetes Metab. Disord. 2019, 18, 51–57. [Google Scholar] [CrossRef]
- Tan, B.; Ma, Y.; Zhang, L.; Li, N.; Zhang, J. The application of metabolomics analysis in the research of gestational diabetes mellitus and preeclampsia. J. Obs. Gynaecol. Res. 2020, 46, 1310–1318. [Google Scholar] [CrossRef]
- Liu, Y.-K.; Hu, W.-T.; Tang, L.-L.; Sheng, Y.-R.; Wei, C.-Y.; Li, M.-Q.; Zhu, X.-Y. Elevated heme impairs macrophage phagocytosis in endometriosis. Reproduction 2019, 158, 257–266. [Google Scholar] [CrossRef]
- Wang, X.; Lee, C.; Li, R.H.W.; Vijayan, M.; Duan, Y.; Yeung, W.S.B.; Zhang, Y.; Chiu, P.C.N. Alteration of the immune cell profiles in the pathophysiology of tubal ectopic pregnancy. Am. J. Reprod. Immunol. 2019, 81, e13093. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.-Z.; Yang, H.-L.; Shi, J.-W.; Shen, H.-H.; Wang, Y.; Chang, K.-K.; Zhang, T.; Ye, J.-F.; Sun, J.-S.; Qiu, X.-M.; et al. Protopanaxadiol improves endometriosis associated infertility and miscarriage in sex hormones receptors-dependent and independent manners. Int. J. Biol. Sci. 2021, 17, 1878–1894. [Google Scholar] [CrossRef] [PubMed]
- Galaz, J.; Romero, R.; Slutsky, R.; Xu, Y.; Motomura, K.; Para, R.; Pacora, P.; Panaitescu, B.; Hsu, C.-D.; Kacerovsky, M.; et al. Cellular immune responses in amniotic fluid of women with preterm prelabor rupture of membranes. J. Perinat. Med. 2020, 48, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Escobar, M.F.; Hincapie, M.A.; Barona, J.S. Immunological Role of the Maternal Uterine Microbiota in Postpartum Hemorrhage. Front. Immunol. 2020, 11, 504. [Google Scholar] [CrossRef] [PubMed]
- Negishi, Y.; Shima, Y.; Kato, M.; Ichikawa, T.; Ino, H.; Horii, Y.; Suzuki, S.; Morita, R. Inflammation in preterm birth: Novel mechanism of preterm birth associated with innate and acquired immunity. J. Reprod. Immunol. 2022, 154, 103748. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.; Timpson, N.; Davey Smith, G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.B.; Kurki, M.I.; Foley, C.N.; Mechakra, A.; Chen, C.-Y.; Marshall, E.; Wilk, J.B.; Ghen, C.-Y.; Runz, H.; Chahine, M.; et al. Genetic associations of protein-coding variants in human disease. Nature 2022, 603, 95–102. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Fauman, E.B.; Petersen, A.-K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte, I.; Forgetta, V.; Yang, T.P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef]
- Krumsiek, J.; Suhre, K.; Evans, A.M.; Mitchell, M.W.; Mohney, R.P.; Milburn, M.V.; Wägele, B.; Römisch-Margl, W.; Illig, T.; Adamski, J.; et al. Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information. PLoS Genet. 2012, 8, e1003005. [Google Scholar] [CrossRef]
- Suhre, K.; Shin, S.Y.; Petersen, A.K.; Mohney, R.P.; Meredith, D.; Wagele, B.; Altmaier, E.; Gram, C.; Deloukas, P.; Erdmann, J.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477, 54–60. [Google Scholar] [CrossRef]
- International HapMap Consortium. A second-generation human haplotype map of over 3.1 million SNPs. Nature 2007, 449, 851–861. [Google Scholar] [CrossRef]
- Orrù, V.; Steri, M.; Sidore, C.; Marongiu, M.; Serra, V.; Olla, S.; Sole, G.; Lai, S.; Dei, M.; Mulas, A.; et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat. Genet. 2020, 52, 1036–1045. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, D.; Zhang, D.; Zuo, X.; Yao, L.; Liu, T.; Ge, X.; He, C.; Zhou, Y.; Shen, Z. Causal role of immune cells in schizophrenia: Mendelian randomization (MR) study. BMC Psychiatry 2023, 23, 590. [Google Scholar] [CrossRef]
- Sidore, C.; Busonero, F.; Maschio, A.; Porcu, E.; Naitza, S.; Zoledziewska, M.; Mulas, A.; Pistis, G.; Steri, M.; Danjou, F.; et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. 2015, 47, 1272–1281. [Google Scholar] [CrossRef]
- Xiang, M.; Wang, Y.; Gao, Z.; Wang, J.; Chen, Q.; Sun, Z.; Liang, J.; Xu, J. Exploring causal correlations between inflammatory cytokines and systemic lupus erythematosus: A Mendelian randomization. Front. Immunol. 2023, 13, 985729. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, R.; Li, C.; Cai, X.; Huo, Z.; Liu, Z.; Ge, F.; Huang, C.; Lu, Y.; Zhong, R.; et al. Causal effects of genetically determined metabolites on cancers included lung, breast, ovarian cancer, and glioma: A Mendelian randomization study. Transl. Lung Cancer Res. 2022, 11, 1302–1314. [Google Scholar] [CrossRef]
- Hemani, G.; Zhengn, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Shim, H.; Chasman, D.I.; Smith, J.D.; Mora, S.; Ridker, P.M.; Nickerson, D.A.; Krauss, R.M.; Stephens, M. A Multivariate Genome-Wide Association Analysis of 10 LDL Subfractions, and Their Response to Statin Treatment, in 1868 Caucasians. PLoS ONE 2015, 10, e0120758. [Google Scholar] [CrossRef] [PubMed]
- Holsinger, K.E.; Weir, B.S. Genetics in geographically structured populations: Defining, estimating and interpreting FST. Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Curtin, F.; Schulz, P. Multiple correlations and Bonferroni’s correction. Biol. Psychiatry 1998, 44, 775–777. [Google Scholar] [CrossRef]
- Bowden, J.; Smith, G.D.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Smith, G.D.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef]
- Sanderson, E.; Spiller, W.; Bowden, J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomization. Stat. Med. 2021, 40, 5434–5452. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Burgess, S.; Davey Smith, G.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Kutalik, Z.; Holmes, M.V.; Minelli, C.; et al. Guidelines for performing Mendelian randomization investigations: Update for summer 2023. Wellcome Open Res. 2023, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Kia, D.A.; Zhang, D.; Guelfi, S.; Manzoni, C.; Hubbard, L.; Reynolds, R.H.; Botía, J.; Ryten, M.; Ferrari, R.; Lewis, P.A.; et al. Identification of Candidate Parkinson Disease Genes by Integrating Genome-Wide Association Study, Expression, and Epigenetic Data Sets. JAMA Neurol. 2021, 78, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.X.; Gloudemans, M.J.; Rao, A.S.; Ingelsson, E.; Montgomery, S.B. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019, 51, 768–769. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Tawfeek, M.A.; Alfadhli, E.M.; Alayoubi, A.M.; El-Beshbishy, H.A.; Habib, F.A. Sex hormone binding globulin as a valuable biochemical marker in predicting gestational diabetes mellitus. BMC Women’s Health 2017, 17, 18. [Google Scholar] [CrossRef]
- Takmaz, T.; Yalvaç, E.S.; Özcan, P.; Çoban, U.; Karasu, A.F.G.; Ünsal, M. The predictive value of weight gain and waist circumference for gestational diabetes mellitus. Turk. J. Obstet. Gynecol. 2019, 16, 199–204. [Google Scholar] [CrossRef]
- Xie, W.H.; Wang, Y.; Xiao, S.Y.; Qiu, L.; Yu, Y.; Zhang, Z.L. Association of gestational diabetes mellitus with overall and type specific cardiovascular and cerebrovascular diseases: Systematic review and meta-analysis. BMJ 2022, 378, e070244. [Google Scholar] [CrossRef]
- Fethney, J. Statistical and clinical significance, and how to use confidence intervals to help interpret both. Aust. Crit. Care 2010, 23, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Li, G.X.; Gao, W.L.; Xu, Y.J.; Xie, M.K.; Tang, S.H.; Yin, P.; Guo, S.; Chu, S.; Sultana, S.; Cui, S. Serum metabonomics study of pregnant women with gestational diabetes mellitus based on LC-MS. Saudi J. Biol. Sci. 2019, 26, 2057–2063. [Google Scholar] [CrossRef]
- Lu, W.Q.; Luo, M.J.; Fang, X.N.; Zhang, R.; Li, S.S.; Tang, M.Y.; Tang, M.; Yu, X.; Hu, C. Discovery of metabolic biomarkers for gestational diabetes mellitus in a Chinese population. Nutr. Metab. 2021, 18, 79. [Google Scholar] [CrossRef]
- Akazawa, S.; Metzger, B.E.; Freinkel, N. Relationships between Glucose and Mannose during Late Gestation in Normal-Pregnancy and Pregnancy Complicated by Diabetes-Mellitus—Concurrent Concentrations in Maternal Plasma and Amniotic-Fluid. J. Clin. Endocr. Metab. 1986, 62, 984–989. [Google Scholar] [CrossRef]
- Feng, D.; Shi, B.; Bi, F.; Sagnelli, M.; Sun, X.; Jiao, J.; Wang, X.; Li, D. Elevated Serum Mannose Levels as a Marker of Polycystic Ovary Syndrome. Front. Endocrinol. 2019, 10, 711. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Stančáková, A.; Lotta, L.A.; Kuusisto, J.; Boren, J.; Blüher, M.; Wareham, N.J.; Ferrannini, E.; Groop, P.H.; Laakso, M.; et al. Plasma Mannose Levels Are Associated with Incident Type 2 Diabetes and Cardiovascular Disease. Cell Metab. 2017, 26, 281–283. [Google Scholar] [CrossRef]
- Ferrannini, E.; Marx, N.; Andreini, D.; Campi, B.; Saba, A.; Gorini, M.; Gorini, M.; Ferrannini, G.; Milzi, A.; Magnoni, M.; et al. Mannose as a biomarker of coronary artery disease: Angiographic evidence and clinical significance. Int. J. Cardiol. 2022, 346, 86–92. [Google Scholar] [CrossRef]
- Lee, S.; Zhang, C.; Kilicarslan, M.; Piening, B.D.; Bjornson, E.; Hallström, B.M.; Groen, A.K.; Ferrannini, E.; Laakso, M.; Snyder, M.; et al. Integrated Network Analysis Reveals an Association between Plasma Mannose Levels and Insulin Resistance. Cell Metab. 2016, 24, 172–184. [Google Scholar] [CrossRef]
- Sharma, V.; Smolin, J.; Nayak, J.; Ayala, J.E.; Scott, D.A.; Peterson, S.N.; Freeze, H.H. Mannose Alters Gut Microbiome, Prevents Diet-Induced Obesity, and Improves Host Metabolism. Cell Rep. 2018, 24, 3087–3098. [Google Scholar] [CrossRef]
- Shin, D.; Lee, K.W.; Song, W.O. Dietary Patterns during Pregnancy Are Associated with Risk of Gestational Diabetes Mellitus. Nutrients 2015, 7, 9369–9382. [Google Scholar] [CrossRef]
- Raimondo, S.; Gentile, M.; Esposito, G.; Gentile, T.; Ferrara, I.; Crescenzo, C.; Palmieri, M.; Cuomo, F.; De Filippo, S.; Lettieri, G.; et al. Could Kallikrein-Related Serine Peptidase 3 Be an Early Biomarker of Environmental Exposure in Young Women? Int. J. Environ. Res. Public Health 2021, 18, 8833. [Google Scholar] [CrossRef]
- Lettieri, G.; Marinaro, C.; Brogna, C.; Montano, L.; Lombardi, M.; Trotta, A.; Troisi, J.; Piscopo, M. A Metabolomic Analysis to Assess the Responses of the Male Gonads of Mytilus galloprovincialis after Heavy Metal Exposure. Metabolites 2023, 13, 1168. [Google Scholar] [CrossRef]
- Lettieri, G.; Marinaro, C.; Notariale, R.; Perrone, P.; Lombardi, M.; Trotta, A.; Troisi, J.; Piscopo, M. Impact of Heavy Metal Exposure on Mytilus galloprovincialis Spermatozoa: A Metabolomic Investigation. Metabolites 2023, 13, 943. [Google Scholar] [CrossRef]
- Hazlina, N.H.N.; Norhayati, M.N.; Bahari, I.S.; Arif, N.A.N.M. Worldwide prevalence, risk factors and psychological impact of infertility among women: A systematic review and meta-analysis. BMJ Open 2022, 12, e057132. [Google Scholar] [CrossRef] [PubMed]
- Shakerian, B.; Irvani, S.; Mostafavi, S.; Moghtaderi, M. Quantitative serum determination of CD3, CD4, CD8, CD16, and CD56 in women with primary infertility: The role of cell-mediated immunity. Turk. J. Obstet. Gynecol. 2022, 19, 242–245. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Mao, X.; Lei, H.; Dong, B.; Guo, D.; Zheng, B.; Sun, P. Peripheral Blood Inflammatory-Immune Cells as a Predictor of Infertility in Women with Polycystic Ovary Syndrome. J. Inflamm. Res. 2020, 13, 441–450. [Google Scholar] [CrossRef]
- Sun, S.; Jiao, M.; Han, C.; Zhang, Q.; Shi, W.; Shi, J.; Li, X. Causal Effects of Genetically Determined Metabolites on Risk of Polycystic Ovary Syndrome: A Mendelian Randomization Study. Front. Endocrinol. 2020, 11, 621. [Google Scholar] [CrossRef]
- de Bennetot, M.; Rabischong, B.; Aublet-Cuvelier, B.; Belard, F.; Fernandez, H.; Bouyer, J.; Canis, M.; Pouly, J.-L. Fertility after tubal ectopic pregnancy: Results of a population-based study. Fertil. Steril. 2012, 98, 1271–1276.e3. [Google Scholar] [CrossRef]
- Shojaei, Z.; Jafarpour, R.; Mehdizadeh, S.; Bayatipoor, H.; Pashangzadeh, S.; Motallebnezhad, M. Functional prominence of natural killer cells and natural killer T cells in pregnancy and infertility: A comprehensive review and update. Pathol. Res. Pract. 2022, 238, 154062. [Google Scholar] [CrossRef]
- Wicherek, L.; Galazka, K.; Lazar, A. Analysis of metallothionein, RCAS1 immunoreactivity regarding immune cell concentration in the endometrium and tubal mucosa in ectopic pregnancy during the course of tubal rupture. Gynecol. Obstet. Investig. 2008, 65, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Daponte, A.; Pournaras, S.; Deligeoroglou, E.; Skentou, H.; Messinis, I.E. Serum interleukin-1beta, interleukin-8 and anti-heat shock 60 Chlamydia trachomatis antibodies as markers of ectopic pregnancy. J. Reprod. Immunol. 2012, 93, 102–108. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Wen, S.; Jia, H.; Du, Y. Evaluation of serum biomarkers and efficacy of MTX in women with ectopic pregnancy. Mol. Med. Rep. 2019, 20, 2902–2908. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, H.-H.; Zhang, Y.-Y.; Wang, X.-Y.; Wang, C.-J.; Wang, Y.; Ye, J.-F.; Li, M.-Q. Potential Causal Association between Plasma Metabolites, Immunophenotypes, and Female Reproductive Disorders: A Two-Sample Mendelian Randomization Analysis. Biomolecules 2024, 14, 116. https://doi.org/10.3390/biom14010116
Shen H-H, Zhang Y-Y, Wang X-Y, Wang C-J, Wang Y, Ye J-F, Li M-Q. Potential Causal Association between Plasma Metabolites, Immunophenotypes, and Female Reproductive Disorders: A Two-Sample Mendelian Randomization Analysis. Biomolecules. 2024; 14(1):116. https://doi.org/10.3390/biom14010116
Chicago/Turabian StyleShen, Hui-Hui, Yang-Yang Zhang, Xuan-Yu Wang, Cheng-Jie Wang, Ying Wang, Jiang-Feng Ye, and Ming-Qing Li. 2024. "Potential Causal Association between Plasma Metabolites, Immunophenotypes, and Female Reproductive Disorders: A Two-Sample Mendelian Randomization Analysis" Biomolecules 14, no. 1: 116. https://doi.org/10.3390/biom14010116




