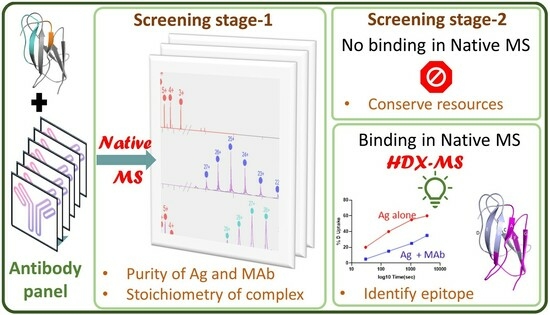
Epitope Mapping of Japanese Encephalitis Virus Neutralizing Antibodies by Native Mass Spectrometry and Hydrogen/Deuterium Exchange
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. MAbs and JEV E-DIII
2.3. Native Mass Spectrometry Analysis
2.4. JEV E-DIII Intact Mass Measurement Using Regular Electrospray Ionization (ESI)
2.5. Hydrogen/Deuterium Exchange Mass Spectrometry
3. Results and Discussion
3.1. Native-MS Analysis of Five MAbs That Bind to Lateral Ridge of JEV E-DIII
3.2. JEV-27 mAb Does Not Bind JEV E-DIII
3.3. Native-MS and HDX-MS for Epitope Mapping
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campbell, G.L.; Hills, S.L.; Fischer, M.; Jacobson, J.A.; Hoke, C.H.; Hombach, J.M.; Marfin, A.A.; Solomon, T.; Tsai, T.F.; Tsu, V.D.; et al. Estimated global incidence of Japanese encephalitis: A systematic review. Bull. World Health Organ 2011, 89, 766–774, 774A–774E. [Google Scholar] [CrossRef]
- Wang, H.; Liang, G. Epidemiology of Japanese encephalitis: Past, present, and future prospects. Ther. Clin. Risk Manag. 2015, 11, 435–448. [Google Scholar] [CrossRef]
- Schuh, A.J.; Ward, M.J.; Leigh Brown, A.J.; Barrett, A.D. Dynamics of the emergence and establishment of a newly dominant genotype of Japanese encephalitis virus throughout Asia. J. Virol. 2014, 88, 4522–4532. [Google Scholar] [CrossRef]
- Plotkin, S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef]
- Plotkin, S.A. Complex correlates of protection after vaccination. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2013, 56, 1458–1465. [Google Scholar] [CrossRef]
- Goncalvez, A.P.; Chien, C.H.; Tubthong, K.; Gorshkova, I.; Roll, C.; Donau, O.; Schuck, P.; Yoksan, S.; Wang, S.D.; Purcell, R.H.; et al. Humanized monoclonal antibodies derived from chimpanzee Fabs protect against Japanese encephalitis virus in vitro and in vivo. J. Virol. 2008, 82, 7009–7021. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaia, N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017, 16, 315–337. [Google Scholar] [CrossRef]
- The Antibody Society. Therapeutic Monoclonal Antibodies Approved or in Regulatory Review. Available online: www.antibodysociety.org/antibody-therapeutics-product-data (accessed on 14 March 2024).
- Barlow, D.J.; Edwards, M.S.; Thornton, J.M. Continuous and discontinuous protein antigenic determinants. Nature 1986, 322, 747–748. [Google Scholar] [CrossRef]
- Toride King, M.; Brooks, C.L. Epitope Mapping of Antibody-Antigen Interactions with X-ray Crystallography. Methods Mol. Biol. 2018, 1785, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Bardelli, M.; Livoti, E.; Simonelli, L.; Pedotti, M.; Moraes, A.; Valente, A.P.; Varani, L. Epitope mapping by solution NMR spectroscopy. J. Mol. Recognit. 2015, 28, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Wigge, C.; Stefanovic, A.; Radjainia, M. The rapidly evolving role of cryo-EM in drug design. Drug Discov. Today Technol. 2020, 38, 91–102. [Google Scholar] [CrossRef]
- Renaud, J.P.; Chung, C.W.; Danielson, U.H.; Egner, U.; Hennig, M.; Hubbard, R.E.; Nar, H. Biophysics in drug discovery: Impact, challenges and opportunities. Nat. Rev. Drug Discov. 2016, 15, 679–698. [Google Scholar] [CrossRef]
- Geysen, H.M.; Meloen, R.H.; Barteling, S.J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. USA 1984, 81, 3998–4002. [Google Scholar] [CrossRef]
- Böttger, V.; Böttger, A. Epitope mapping using phage display peptide libraries. Methods Mol. Biol. 2009, 524, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, B.C.; Wells, J.A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 1989, 244, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.A.; Watanabe, C.K.; Zhong, A.; Goddard, A.; Sidhu, S.S. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. USA 2000, 97, 8950–8954. [Google Scholar] [CrossRef] [PubMed]
- Davidson, E.; Doranz, B.J. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology 2014, 143, 13–20. [Google Scholar] [CrossRef]
- Säfsten, P. Epitope mapping by surface plasmon resonance. Methods Mol. Biol. 2009, 524, 67–76. [Google Scholar] [CrossRef]
- Suckau, D.; Köhl, J.; Karwath, G.; Schneider, K.; Casaretto, M.; Bitter-Suermann, D.; Przybylski, M. Molecular epitope identification by limited proteolysis of an immobilized antigen-antibody complex and mass spectrometric peptide mapping. Proc. Natl. Acad. Sci. USA 1990, 87, 9848–9852. [Google Scholar] [CrossRef]
- Zhao, Y.; Chalt, B.T. Protein epitope mapping by mass spectrometry. Anal. Chem. 1994, 66, 3723–3726. [Google Scholar] [CrossRef]
- Fernandez, E.; Kose, N.; Edeling, M.A.; Adhikari, J.; Sapparapu, G.; Lazarte, S.M.; Nelson, C.A.; Govero, J.; Gross, M.L.; Fremont, D.H.; et al. Mouse and Human Monoclonal Antibodies Protect against Infection by Multiple Genotypes of Japanese Encephalitis Virus. mBio 2018, 9, e00008-18. [Google Scholar] [CrossRef]
- Zhang, Q.; Willison, L.N.; Tripathi, P.; Sathe, S.K.; Roux, K.H.; Emmett, M.R.; Blakney, G.T.; Zhang, H.M.; Marshall, A.G. Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2011, 83, 7129–7136. [Google Scholar] [CrossRef]
- Adhikari, J.; Zhao, H.; Fernandez, E.; Huang, Y.; Diamond, M.S.; Fremont, D.H.; Gross, M.L. Hydrogen-deuterium exchange mass spectrometry identifies spatially distinct antibody epitopes on domain III of the Zika virus envelope protein. J. Mass Spectrom. 2021, 56, e4685. [Google Scholar] [CrossRef]
- Doyle, M.P.; Genualdi, J.R.; Bailey, A.L.; Kose, N.; Gainza, C.; Rodriguez, J.; Reeder, K.M.; Nelson, C.A.; Jethva, P.N.; Sutton, R.E.; et al. Isolation of a Potently Neutralizing and Protective Human Monoclonal Antibody Targeting Yellow Fever Virus. mBio 2022, 13, e0051222. [Google Scholar] [CrossRef]
- Zhang, Y.; Wecksler, A.T.; Molina, P.; Deperalta, G.; Gross, M.L. Mapping the Binding Interface of VEGF and a Monoclonal Antibody Fab-1 Fragment with Fast Photochemical Oxidation of Proteins (FPOP) and Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.M.; Sperry, J.B.; Carroll, J.A.; Gross, M.L. Fast photochemical oxidation of proteins for epitope mapping. Anal. Chem. 2011, 83, 7657–7661. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Chance, M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007, 107, 3514–3543. [Google Scholar] [CrossRef] [PubMed]
- Kiselar, J.G.; Chance, M.R. Future directions of structural mass spectrometry using hydroxyl radical footprinting. J. Mass Spectrom. 2010, 45, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Pimenova, T.; Nazabal, A.; Roschitzki, B.; Seebacher, J.; Rinner, O.; Zenobi, R. Epitope mapping on bovine prion protein using chemical cross-linking and mass spectrometry. J. Mass Spectrom. 2008, 43, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Lossl, P.; Sinz, A. Combining Amine-Reactive Cross-Linkers and Photo-Reactive Amino Acids for 3D-Structure Analysis of Proteins and Protein Complexes. Methods Mol. Biol. 2016, 1394, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Huang, R.Y.; Beno, B.R.; Deyanova, E.G.; Li, J.; Chen, G.; Gross, M.L. Epitope and Paratope Mapping of PD-1/Nivolumab by Mass Spectrometry-Based Hydrogen-Deuterium Exchange, Cross-linking, and Molecular Docking. Anal. Chem. 2020, 92, 9086–9094. [Google Scholar] [CrossRef] [PubMed]
- Rosati, S.; Yang, Y.; Barendregt, A.; Heck, A.J. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat. Protoc. 2014, 9, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Leney, A.C.; Heck, A.J. Native Mass Spectrometry: What is in the Name? J. Am. Soc. Mass Spectrom. 2017, 28, 5–13. [Google Scholar] [CrossRef]
- Terral, G.; Champion, T.; Debaene, F.; Colas, O.; Bourguet, M.; Wagner-Rousset, E.; Corvaia, N.; Beck, A.; Cianferani, S. Epitope characterization of anti-JAM-A antibodies using orthogonal mass spectrometry and surface plasmon resonance approaches. MAbs 2017, 9, 1317–1326. [Google Scholar] [CrossRef]
- Zhang, Y.; Cui, W.; Wecksler, A.T.; Zhang, H.; Molina, P.; Deperalta, G.; Gross, M.L. Native MS and ECD Characterization of a Fab-Antigen Complex May Facilitate Crystallization for X-ray Diffraction. J. Am. Soc. Mass Spectrom. 2016, 27, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Tito, M.A.; Miller, J.; Walker, N.; Griffin, K.F.; Williamson, E.D.; Despeyroux-Hill, D.; Titball, R.W.; Robinson, C.V. Probing molecular interactions in intact antibody: Antigen complexes, an electrospray time-of-flight mass spectrometry approach. Biophys. J. 2001, 81, 3503–3509. [Google Scholar] [CrossRef]
- Atmanene, C.; Wagner-Rousset, E.; Malissard, M.; Chol, B.; Robert, A.; Corvaïa, N.; Van Dorsselaer, A.; Beck, A.; Sanglier-Cianférani, S. Extending mass spectrometry contribution to therapeutic monoclonal antibody lead optimization: Characterization of immune complexes using noncovalent ESI-MS. Anal. Chem. 2009, 81, 6364–6373. [Google Scholar] [CrossRef]
- Chalmers, M.J.; Busby, S.A.; Pascal, B.D.; He, Y.; Hendrickson, C.L.; Marshall, A.G.; Griffin, P.R. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 2006, 78, 1005–1014. [Google Scholar] [CrossRef]
- Rehder, D.S.; Chelius, D.; McAuley, A.; Dillon, T.M.; Xiao, G.; Crouse-Zeineddini, J.; Vardanyan, L.; Perico, N.; Mukku, V.; Brems, D.N.; et al. Isomerization of a single aspartyl residue of anti-epidermal growth factor receptor immunoglobulin gamma2 antibody highlights the role avidity plays in antibody activity. Biochemistry 2008, 47, 2518–2530. [Google Scholar] [CrossRef]
- Tian, Y.; Ruotolo, B.T. The growing role of structural mass spectrometry in the discovery and development of therapeutic antibodies. Analyst 2018, 143, 2459–2468. [Google Scholar] [CrossRef]
- Thompson, N.J.; Hendriks, L.J.; de Kruif, J.; Throsby, M.; Heck, A.J. Complex mixtures of antibodies generated from a single production qualitatively and quantitatively evaluated by native Orbitrap mass spectrometry. MAbs 2014, 6, 197–203. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Vimer, S.; Warszawski, S.; Katz, A.; Yona, M.; Unger, T.; Peleg, Y.; Morgenstern, D.; Cohen-Dvashi, H.; Diskin, R.; et al. Rapid characterization of secreted recombinant proteins by native mass spectrometry. Commun. Biol. 2018, 1, 213. [Google Scholar] [CrossRef]
- Yefremova, Y.; Opuni, K.F.M.; Danquah, B.D.; Thiesen, H.J.; Glocker, M.O. Intact Transition Epitope Mapping (ITEM). J. Am. Soc. Mass Spectrom. 2017, 28, 1612–1622. [Google Scholar] [CrossRef]
- Jethva, P.N.; Gross, M.L. Hydrogen deuterium exchange and other mass spectrometry- based approaches for epitope mapping. Front. Anal. Sci. 2023, 3, 1118749. [Google Scholar] [CrossRef]
- Huang, Y.; Salinas, N.D.; Chen, E.; Tolia, N.H.; Gross, M.L. Native Mass Spectrometry, Ion mobility, and Collision-Induced Unfolding Categorize Malaria Antigen/Antibody Binding. J. Am. Soc. Mass Spectrom. 2017, 28, 2515–2518. [Google Scholar] [CrossRef]
- den Boer, M.A.; Lai, S.-H.; Xue, X.; van Kampen, M.D.; Bleijlevens, B.; Heck, A.J.R. Comparative Analysis of Antibodies and Heavily Glycosylated Macromolecular Immune Complexes by Size-Exclusion Chromatography Multi-Angle Light Scattering, Native Charge Detection Mass Spectrometry, and Mass Photometry. Anal. Chem. 2022, 94, 892–900. [Google Scholar] [CrossRef]
- Zhang, Q.; Noble, K.A.; Mao, Y.; Young, N.L.; Sathe, S.K.; Roux, K.H.; Marshall, A.G. Rapid screening for potential epitopes reactive with a polycolonal antibody by solution-phase H/D exchange monitored by FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 2013, 24, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Dowd, K.A.; Brien, J.D.; Edeling, M.A.; Gorlatov, S.; Johnson, S.; Lee, I.; Akahata, W.; Nabel, G.J.; Richter, M.K.; et al. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 2013, 9, e1003312. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Fernandez, E.; Dowd, K.A.; Speer, S.D.; Platt, D.J.; Gorman, M.J.; Govero, J.; Nelson, C.A.; Pierson, T.C.; Diamond, M.S. Structural basis of Zika virus-specific antibody protection. Cell 2016, 166, 1016–1027. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, I.D.; Robinson, J.H.; Hui, J.O.; Shi, S.D.-H.; Netirojjanakul, C.; Nshanian, M.; Egea, P.F.; Lippens, J.L.; Bagal, D.; Loo, J.A. Native and denaturing MS protein deconvolution for biopharma: Monoclonal antibodies and antibody–drug conjugates to polydisperse membrane proteins and beyond. Anal. Chem. 2019, 91, 9472–9480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Marshall, A.G. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998, 9, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Wu, S.C. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 2003, 77, 2600–2606. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Fox, J.M.; Hawman, D.W.; Huang, Y.J.; Messaoudi, I.; Kreklywich, C.; Denton, M.; Legasse, A.W.; Smith, P.P.; Johnson, S.; et al. Chikungunya viruses that escape monoclonal antibody therapy are clinically attenuated, stable, and not purified in mosquitoes. J. Virol. 2014, 88, 8213–8226. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.A. Electrospray ionization mass spectrometry: A technology for studying noncovalent macromolecular complexes. Int. J. Mass Spectrom. 2000, 200, 175–186. [Google Scholar] [CrossRef]
- Luca, V.C.; AbiMansour, J.; Nelson, C.A.; Fremont, D.H. Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 2012, 86, 2337–2346. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adhikari, J.; Heffernan, J.; Edeling, M.; Fernandez, E.; Jethva, P.N.; Diamond, M.S.; Fremont, D.H.; Gross, M.L. Epitope Mapping of Japanese Encephalitis Virus Neutralizing Antibodies by Native Mass Spectrometry and Hydrogen/Deuterium Exchange. Biomolecules 2024, 14, 374. https://doi.org/10.3390/biom14030374
Adhikari J, Heffernan J, Edeling M, Fernandez E, Jethva PN, Diamond MS, Fremont DH, Gross ML. Epitope Mapping of Japanese Encephalitis Virus Neutralizing Antibodies by Native Mass Spectrometry and Hydrogen/Deuterium Exchange. Biomolecules. 2024; 14(3):374. https://doi.org/10.3390/biom14030374
Chicago/Turabian StyleAdhikari, Jagat, James Heffernan, Melissa Edeling, Estefania Fernandez, Prashant N. Jethva, Michael S. Diamond, Daved H. Fremont, and Michael L. Gross. 2024. "Epitope Mapping of Japanese Encephalitis Virus Neutralizing Antibodies by Native Mass Spectrometry and Hydrogen/Deuterium Exchange" Biomolecules 14, no. 3: 374. https://doi.org/10.3390/biom14030374