Technical Advances in Circulating Cell-Free DNA Detection and Analysis for Personalized Medicine in Patients’ Care
Abstract
1. Introduction
2. Circulating Cell-Free DNA
3. cfDNA Applications in Clinical Care
3.1. Oncologic Applications
3.2. Prenatal Screening
3.3. Transplantation
4. Technical Issues for High-Quality cfDNA Analysis
4.1. The Relevance of Correct Sampling
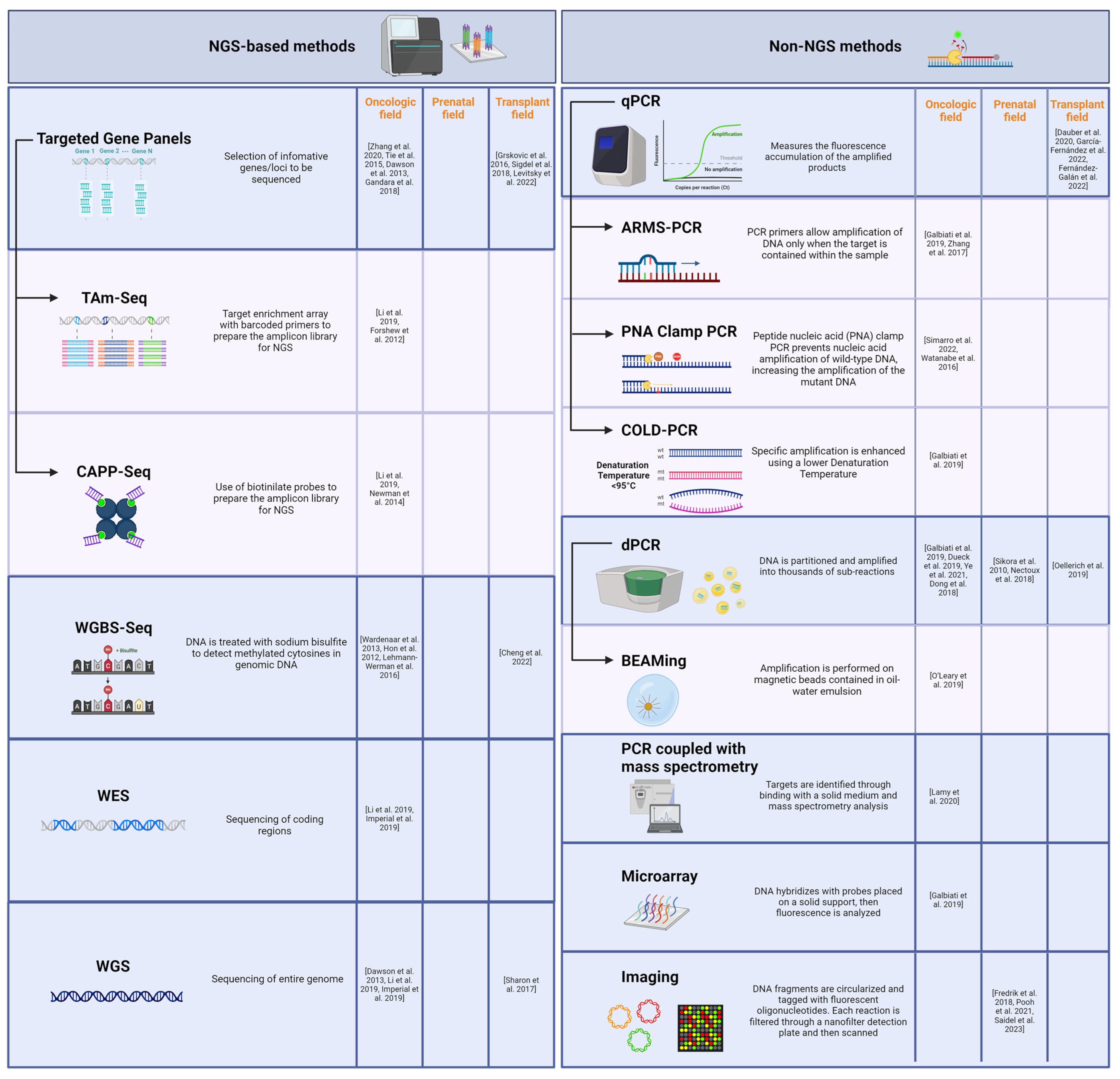
4.2. Technical Comparison of cfDNA Analysis Methods
4.2.1. NGS-Based Methods
4.2.2. Non-NGS Methods
5. Conclusions
Funding
Conflicts of Interest
References
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372, eaaw3616. [Google Scholar] [CrossRef] [PubMed]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jürchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar] [PubMed]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature: The diverse origins of circulating cell-free DNA. Biol. Rev. 2018, 93, 1649–1683. [Google Scholar] [CrossRef] [PubMed]
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rosales Rodriguez, I.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020, 21, 8062. [Google Scholar] [CrossRef] [PubMed]
- McCoubrey-Hoyer, A.; Okarma, T.B.; Holman, H.R. Partial purification and characterization of plasma DNA and its relation to disease activity in systemic lupus erythematosus. Am. J. Med. 1984, 77, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Chan, K.C.A.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.F.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef] [PubMed]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Rumore, P.M.; Steinman, C.R. Endogenous circulating DNA in systemic lupus erythematosus. Occurrence as multimeric complexes bound to histone. J. Clin. Investig. 1990, 86, 69–74. [Google Scholar] [CrossRef]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-free DNA in human blood plasma: Length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef]
- Edwards, R.L.; Menteer, J.; Lestz, R.M.; Baxter-Lowe, L.A. Cell-free DNA as a solid-organ transplant biomarker: Technologies and approaches. Biomark. Med. 2022, 16, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Moreira, V.G.; Prieto, B.; Rodríguez, J.S.M.; Alvarez, F.V. Usefulness of cell-free plasma DNA, procalcitonin and C-reactive protein as markers of infection in febrile patients. Ann. Clin. Biochem. 2010, 47 Pt 3, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Burnham, P.; Dadhania, D.; Heyang, M.; Chen, F.; Westblade, L.F.; Suthanthiran, M.; Lee, J.R.; De Vlaminck, I. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 2018, 9, 2412. [Google Scholar] [CrossRef] [PubMed]
- Siljan, W.W.; Holter, J.C.; Nymo, S.H.; Schjalm, C.; Müller, F.; Jenum, P.A.; Husebye, E.; Ueland, T.; Aukrust, P.; Mollnes, T.E.; et al. Circulating cell-free DNA is elevated in community-acquired bacterial pneumonia and predicts short-term outcome. J. Infect. 2016, 73, 383–386. [Google Scholar] [CrossRef]
- Cisneros-Villanueva, M.; Hidalgo-Pérez, L.; Rios-Romero, M.; Cedro-Tanda, A.; Ruiz-Villavicencio, C.A.; Page, K.; Hastings, R.; Fernandez-Garcia, D.; Allsopp, R.; Fonseca-Montaño, M.A.; et al. Cell-free DNA analysis in current cancer clinical trials: A review. Br. J. Cancer 2022, 126, 391–400. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, H.M.; Feber, A.; Popat, S. Minimal Residual Disease Monitoring in Radically Treated Non-Small Cell Lung Cancer: Challenges and Future Directions. OncoTargets Ther. 2023, 16, 249–259. [Google Scholar] [CrossRef]
- Spigel, D.R.; Ervin, T.J.; Ramlau, R.; Daniel, D.B.; Goldschmidt, J.H.; Blumenschein, G.R.; Krzakowski, M.J.; Robinet, G.; Clement-Duchene, C.; Barlesi, F.; et al. Final efficacy results from OAM4558g, a randomized phase II study evaluating MetMAb or placebo in combination with erlotinib in advanced NSCLC. J. Clin. Oncol. 2011, 29 (Suppl. S15), 7505. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Y.-L.; Chen, G.; Feng, J.; Liu, X.-Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef]
- Gatzemeier, U.; Pluzanska, A.; Szczesna, A.; Kaukel, E.; Roubec, J.; Brennscheidt, U.; De Rosa, F.; Mueller, B.; Von Pawel, J. Results of a phase III trial of erlotinib (OSI-774) combined with cisplatin and gemcitabine (GC) chemotherapy in advanced non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2004, 22 (Suppl. S14), 7010. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.-H.; Barrett, J.C.; Jänne, P.A. Association Between Plasma Genotyping and Outcomes of Treatment with Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Luft, A.; Vicente, D.; Tafreshi, A.; Gümüş, M.; Mazières, J.; Hermes, B.; Çay Şenler, F.; Csőszi, T.; Fülöp, A.; et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2040–2051. [Google Scholar] [CrossRef]
- Fabrizio, D.; Lieber, D.; Malboeuf, C.; Silterra, J.; White, E.; Coyne, M.; Brennan, T.; Ma, J.; Kennedy, M.; Schleifman, E.; et al. Abstract 5706: A blood-based next-generation sequencing assay to determine tumor mutational burden (bTMB) is associated with benefit to an anti-PD-L1 inhibitor, atezolizumab. Cancer Res. 2018, 78, 5706. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Nathanson, T.; Rizvi, H.; Creelan, B.C.; Sanchez-Vega, F.; Ahuja, A.; Ni, A.; Novik, J.B.; Mangarin, L.M.B.; Abu-Akeel, M.; et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018, 33, 843–852.e4. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Li, B.; Adashek, J.J.; Cha, S.W.; Bianchi-Frias, D.; Qian, D.; Kim, L.; So, T.W.; Mitchell, M.; Kamei, N.; et al. Serial changes in liquid biopsy-derived variant allele frequency predict immune checkpoint inhibitor responsiveness in the pan-cancer setting. Oncoimmunology 2022, 11, 2052410. [Google Scholar] [CrossRef]
- Zhang, Q.; Luo, J.; Wu, S.; Si, H.; Gao, C.; Xu, W.; Abdullah, S.E.; Higgs, B.W.; Dennis, P.A.; van der Heijden, M.S.; et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov. 2020, 10, 1842–1853. [Google Scholar] [CrossRef]
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Swigart, L.B.; Wu, H.-T.; Hirst, G.L.; Yau, C.; Wolf, D.M.; Tin, A.; Salari, R.; Shchegrova, S.; Pawar, H.; et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. 2021, 32, 229–239. [Google Scholar] [CrossRef]
- Cao, H.; Liu, X.; Chen, Y.; Yang, P.; Huang, T.; Song, L.; Xu, R. Circulating Tumor DNA Is Capable of Monitoring the Therapeutic Response and Resistance in Advanced Colorectal Cancer Patients Undergoing Combined Target and Chemotherapy. Front. Oncol. 2020, 10, 466. [Google Scholar] [CrossRef]
- Ma, F.; Zhu, W.; Guan, Y.; Yang, L.; Xia, X.; Chen, S.; Li, Q.; Guan, X.; Yi, Z.; Qian, H.; et al. ctDNA dynamics: A novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget 2016, 7, 66020–66031. [Google Scholar] [CrossRef]
- Dawson, S.-J.; Tsui, D.W.Y.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgård, B.; Blennow, K.; Zetterberg, H.; et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, A.; Liu, M.C.; Klein, E.A.; Venn, O.; Hubbell, E.; Beausang, J.F.; Gross, S.; Melton, C.; Fields, A.P.; Liu, Q.; et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 2022, 40, 1537–1549.e12. [Google Scholar] [CrossRef] [PubMed]
- Rather, R.A.; Saha, S.C. Reappraisal of evolving methods in non-invasive prenatal screening: Discovery, biology and clinical utility. Heliyon 2023, 9, e13923. [Google Scholar] [CrossRef]
- Wright, C.F.; Burton, H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum. Reprod. Update 2009, 15, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Nectoux, J. Current, Emerging, and Future Applications of Digital PCR in Non-Invasive Prenatal Diagnosis. Mol. Diagn. Ther. 2018, 22, 139–148. [Google Scholar] [CrossRef]
- Hui, L.; Bianchi, D.W. Fetal fraction and noninvasive prenatal testing: What clinicians need to know. Prenat. Diagn. 2020, 40, 155–163. [Google Scholar] [CrossRef]
- Gil, M.M.; Accurti, V.; Santacruz, B.; Plana, M.N.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2017, 50, 302–314. [Google Scholar] [CrossRef] [PubMed]
- De Vlaminck, I.; Valantine, H.A.; Snyder, T.M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; Bernstein, D.; Weisshaar, D.; et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med. 2014, 6, 241ra77. [Google Scholar] [CrossRef] [PubMed]
- De Vlaminck, I.; Martin, L.; Kertesz, M.; Patel, K.; Kowarsky, M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. USA 2015, 112, 13336–13341. [Google Scholar] [CrossRef] [PubMed]
- Gielis, E.M.; Ledeganck, K.J.; Dendooven, A.; Meysman, P.; Beirnaert, C.; Laukens, K.; De Schrijver, J.; Van Laecke, S.; Van Biesen, W.; Emonds, M.-P.; et al. The use of plasma donor-derived, cell-free DNA to monitor acute rejection after kidney transplantation. Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transpl. Assoc.-Eur. Ren. Assoc. 2020, 35, 714–721. [Google Scholar] [CrossRef]
- Oellerich, M.; Budde, K.; Osmanodja, B.; Bornemann-Kolatzki, K.; Beck, J.; Schütz, E.; Walson, P.D. Donor-derived cell-free DNA as a diagnostic tool in transplantation. Front. Genet. 2022, 13, 1031894. [Google Scholar] [CrossRef] [PubMed]
- Grskovic, M.; Hiller, D.J.; Eubank, L.A.; Sninsky, J.J.; Christopherson, C.; Collins, J.P.; Thompson, K.; Song, M.; Wang, Y.S.; Ross, D.; et al. Validation of a Clinical-Grade Assay to Measure Donor-Derived Cell-Free DNA in Solid Organ Transplant Recipients. J. Mol. Diagn. JMD 2016, 18, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Archila, F.A.; Constantin, T.; Prins, S.A.; Liberto, J.; Damm, I.; Towfighi, P.; Navarro, S.; Kirkizlar, E.; Demko, Z.P.; et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. J. Clin. Med. 2018, 8, 19. [Google Scholar] [CrossRef]
- Sorbini, M.; Togliatto, G.M.; Simonato, E.; Boffini, M.; Cappuccio, M.; Gambella, A.; Arruga, F.; Mora, N.; Marro, M.; Caorsi, C.; et al. HLA-DRB1 mismatch-based identification of donor-derived cell free DNA (dd-cfDNA) as a marker of rejection in heart transplant recipients: A single-institution pilot study. J. Heart Lung Transplant. Off. Publ. Int. Soc. Heart Transplant. 2021, 40, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Sorbini, M.; Togliatto, G.; Mioli, F.; Simonato, E.; Marro, M.; Cappuccio, M.; Arruga, F.; Caorsi, C.; Mansouri, M.; Magistroni, P.; et al. Validation of a Simple, Rapid, and Cost-Effective Method for Acute Rejection Monitoring in Lung Transplant Recipients. Transpl. Int. Off. J. Eur. Soc. Organ Transplant. 2022, 35, 10546. [Google Scholar] [CrossRef]
- Snyder, T.M.; Khush, K.K.; Valantine, H.A.; Quake, S.R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. USA 2011, 108, 6229–6234. [Google Scholar] [CrossRef]
- Sharon, E.; Shi, H.; Kharbanda, S.; Koh, W.; Martin, L.R.; Khush, K.K.; Valantine, H.; Pritchard, J.K.; De Vlaminck, I. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput. Biol. 2017, 13, e1005629. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-B.; Cutler, C.S. Biomarkers for acute GVHD: Can we predict the unpredictable? Bone Marrow Transplant. 2013, 48, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, M.; Pennisi, S.; Pfeifer, D.; Deuter, M.; von Bubnoff, N.; Scherer, F.; Strüssmann, T.; Wehr, C.; Duyster, J.; Bertz, H.; et al. Colon and liver tissue damage detection using methylated SESN3 and PTK2B genes in circulating cell-free DNA in patients with acute graft-versus-host disease. Bone Marrow Transplant. 2021, 56, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jiang, P.; Chan, K.C.A.; Wong, J.; Cheng, Y.K.Y.; Liang, R.H.S.; Chan, W.; Ma, E.S.K.; Chan, S.L.; Cheng, S.H.; et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 2015, 112, E5503–E5512. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.P.; Cheng, M.P.; Loy, C.J.; Lenz, J.S.; Chen, K.; Smalling, S.; Burnham, P.; Timblin, K.M.; Orejas, J.L.; Silverman, E.; et al. Cell-free DNA profiling informs all major complications of hematopoietic cell transplantation. Proc. Natl. Acad. Sci. USA 2022, 119, e2113476118. [Google Scholar] [CrossRef]
- Levitsky, J.; Kandpal, M.; Guo, K.; Kleiboeker, S.; Sinha, R.; Abecassis, M. Donor-derived cell-free DNA levels predict graft injury in liver transplant recipients. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2022, 22, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, J.G.H.P.; Boer, K.; Peeters, A.M.A.; Clahsen-van Groningen, M.C.; Roodnat, J.I.; van de Wetering, J.; Nieboer, D.; Bost, D.A.; Baan, C.C.; Hesselink, D.A. A Novel High-throughput Droplet Digital PCR-based Indel Quantification Method for the Detection of Circulating Donor-derived Cell-free DNA After Kidney Transplantation. Transplantation 2022, 106, 1777–1786. [Google Scholar] [CrossRef]
- Norton, S.E.; Lechner, J.M.; Williams, T.; Fernando, M.R. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin. Biochem. 2013, 46, 1561–1565. [Google Scholar] [CrossRef]
- Knüttgen, F.; Beck, J.; Dittrich, M.; Oellerich, M.; Zittermann, A.; Schulz, U.; Fuchs, U.; Knabbe, C.; Schütz, E.; Gummert, J.; et al. Graft-derived Cell-free DNA as a Noninvasive Biomarker of Cardiac Allograft Rejection: A Cohort Study on Clinical Validity and Confounding Factors. Transplantation 2022, 106, 615–622. [Google Scholar] [CrossRef]
- Clausen, F.B.; Jørgensen, K.M.C.L.; Wardil, L.W.; Nielsen, L.K.; Krog, G.R. Droplet digital PCR-based testing for donor-derived cell-free DNA in transplanted patients as noninvasive marker of allograft health: Methodological aspects. PLoS ONE 2023, 18, e0282332. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jing, C.; Wu, J.; Ni, J.; Sha, H.; Xu, X.; Du, Y.; Lou, R.; Dong, S.; Feng, J. Circulating tumor DNA detection: A potential tool for colorectal cancer management. Oncol. Lett. 2019, 17, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Wardenaar, R.; Liu, H.; Colot, V.; Colomé-Tatché, M.; Johannes, F. Evaluation of MeDIP-chip in the context of whole-genome bisulfite sequencing (WGBS-seq) in Arabidopsis. Methods Mol. Biol. Clifton NJ 2013, 1067, 203–224. [Google Scholar] [CrossRef]
- Hon, G.C.; Hawkins, R.D.; Caballero, O.L.; Lo, C.; Lister, R.; Pelizzola, M.; Valsesia, A.; Ye, Z.; Kuan, S.; Edsall, L.E.; et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012, 22, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Imperial, R.; Nazer, M.; Ahmed, Z.; Kam, A.E.; Pluard, T.J.; Bahaj, W.; Levy, M.; Kuzel, T.M.; Hayden, D.M.; Pappas, S.G.; et al. Matched Whole-Genome Sequencing (WGS) and Whole-Exome Sequencing (WES) of Tumor Tissue with Circulating Tumor DNA (ctDNA) Analysis: Complementary Modalities in Clinical Practice. Cancers 2019, 11, 1399. [Google Scholar] [CrossRef]
- Dauber, E.-M.; Kollmann, D.; Kozakowski, N.; Rasoul-Rockenschaub, S.; Soliman, T.; Berlakovich, G.A.; Mayr, W.R. Quantitative PCR of INDELs to measure donor-derived cell-free DNA-a potential method to detect acute rejection in kidney transplantation: A pilot study. Transpl. Int. Off. J. Eur. Soc. Organ Transplant. 2020, 33, 298–309. [Google Scholar] [CrossRef]
- García-Fernández, N.; Macher, H.C.; Suárez-Artacho, G.; Gómez-Bravo, M.Á.; Molinero, P.; Guerrero, J.M.; Porras-López, M.; Rubio, A. Donor-Specific Cell-Free DNA qPCR Quantification as a Noninvasive Accurate Biomarker for Early Rejection Detection in Liver Transplantation. J. Clin. Med. 2022, 12, 36. [Google Scholar] [CrossRef]
- Fernández-Galán, E.; Badenas, C.; Fondevila, C.; Jiménez, W.; Navasa, M.; Puig-Butillé, J.A.; Brunet, M. Monitoring of Donor-Derived Cell-Free DNA by Short Tandem Repeats: Concentration of Total Cell-Free DNA and Fragment Size for Acute Rejection Risk Assessment in Liver Transplantation. Liver Transplant. Off. Publ. Am. Assoc. Study Liver Dis. Int. Liver Transplant. Soc. 2022, 28, 257–268. [Google Scholar] [CrossRef]
- Galbiati, S.; Damin, F.; Burgio, V.; Brisci, A.; Soriani, N.; Belcastro, B.; Di Resta, C.; Gianni, L.; Chiari, M.; Ronzoni, M.; et al. Evaluation of three advanced methodologies, COLD-PCR, microarray and ddPCR, for identifying the mutational status by liquid biopsies in metastatic colorectal cancer patients. Clin. Chim. Acta 2019, 489, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chang, N.; Yang, G.; Zhang, Y.; Ye, M.; Cao, J.; Xiong, J.; Han, Z.; Wu, S.; Shang, L.; et al. A comparison of ARMS-Plus and droplet digital PCR for detecting EGFR activating mutations in plasma. Oncotarget 2017, 8, 112014–112023. [Google Scholar] [CrossRef] [PubMed]
- Simarro, J.; Pérez-Simó, G.; Mancheño, N.; Ansotegui, E.; Muñoz-Núñez, C.F.; Gómez-Codina, J.; Juan, Ó.; Palanca, S. Technical Validation and Clinical Implications of Ultrasensitive PCR Approaches for EGFR-Thr790Met Mutation Detection in Pretreatment FFPE Samples and in Liquid Biopsies from Non-Small Cell Lung Cancer Patients. Int. J. Mol. Sci. 2022, 23, 8526. [Google Scholar] [CrossRef]
- Watanabe, K.; Fukuhara, T.; Tsukita, Y.; Morita, M.; Suzuki, A.; Tanaka, N.; Terasaki, H.; Nukiwa, T.; Maemondo, M. EGFR Mutation Analysis of Circulating Tumor DNA Using an Improved PNA-LNA PCR Clamp Method. Can. Respir. J. 2016, 2016, e5297329. [Google Scholar] [CrossRef] [PubMed]
- Lamy, P.-J.; van der Leest, P.; Lozano, N.; Becht, C.; Duboeuf, F.; Groen, H.J.M.; Hilgers, W.; Pourel, N.; Rifaela, N.; Schuuring, E.; et al. Mass Spectrometry as a Highly Sensitive Method for Specific Circulating Tumor DNA Analysis in NSCLC: A Comparison Study. Cancers 2020, 12, 3002. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Hrebien, S.; Beaney, M.; Fribbens, C.; Garcia-Murillas, I.; Jiang, J.; Li, Y.; Huang Bartlett, C.; André, F.; Loibl, S.; et al. Comparison of BEAMing and Droplet Digital PCR for Circulating Tumor DNA Analysis. Clin. Chem. 2019, 65, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Dueck, M.E.; Lin, R.; Zayac, A.; Gallagher, S.; Chao, A.K.; Jiang, L.; Datwani, S.S.; Hung, P.; Stieglitz, E. Precision cancer monitoring using a novel, fully integrated, microfluidic array partitioning digital PCR platform. Sci. Rep. 2019, 9, 19606. [Google Scholar] [CrossRef]
- Sikora, A.; Zimmermann, B.G.; Rusterholz, C.; Birri, D.; Kolla, V.; Lapaire, O.; Hoesli, I.; Kiefer, V.; Jackson, L.; Hahn, S. Detection of increased amounts of cell-free fetal DNA with short PCR amplicons. Clin. Chem. 2010, 56, 136–138. [Google Scholar] [CrossRef]
- Ye, P.; Cai, P.; Xie, J.; Wei, Y. The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0248775. [Google Scholar] [CrossRef]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci. Rep. 2018, 8, 9650. [Google Scholar] [CrossRef]
- Oellerich, M.; Shipkova, M.; Asendorf, T.; Walson, P.D.; Schauerte, V.; Mettenmeyer, N.; Kabakchiev, M.; Hasche, G.; Gröne, H.-J.; Friede, T.; et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2019, 19, 3087–3099. [Google Scholar] [CrossRef] [PubMed]
- Dahl, F.; Ericsson, O.; Karlberg, O.; Karlsson, F.; Howell, M.; Persson, F.; Roos, F.; Stenberg, J.; Ahola, T.; Alftrén, I.; et al. Imaging Single DNA Molecules for High Precision NIPT. Sci. Rep. 2018, 8, 4549. [Google Scholar] [CrossRef] [PubMed]
- Pooh, R.K.; Masuda, C.; Matsushika, R.; Machida, M.; Nakamura, T.; Takeda, M.; Ohashi, H.; Kumagai, M.; Uenishi, K.; Roos, F.; et al. Clinical Validation of Fetal cfDNA Analysis Using Rolling-Circle-Replication and Imaging Technology in Osaka (CRITO Study). Diagnostics 2021, 11, 1837. [Google Scholar] [CrossRef] [PubMed]
- Saidel, M.L.; Ananth, U.; Rose, D.; Farrell, C. Non-Invasive prenatal testing with rolling circle amplification: Real-world clinical experience in a non-molecular laboratory. J. Clin. Lab. Anal. 2023, 37, e24870. [Google Scholar] [CrossRef] [PubMed]
- Laver, T.; Harrison, J.; O’Neill, P.A.; Moore, K.; Farbos, A.; Paszkiewicz, K.; Studholme, D.J. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 2015, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.O.; Russ, C.; Ross, M.G.; Gabriel, S.B.; Nusbaum, C.; DePristo, M.A. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genom. 2012, 13, 375. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Reeve, J.; Madill-Thomsen, K.S.; Demko, Z.; Prewett, A.; Gauthier, P.; Billings, P.; Lawrence, C.; Lowe, D.; Hidalgo, L.G.; et al. Antibody-mediated Rejection without Detectable Donor-specific Antibody Releases Donor-derived Cell-free DNA: Results From the Trifecta Study. Transplantation 2023, 107, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.-M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-throughput sequencing technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef]
- Levy, S.E.; Boone, B.E. Next-Generation Sequencing Strategies. Cold Spring Harb. Perspect. Med. 2019, 9, a025791. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, U.I.; Gulilat, M.; Kim, R.B. The Role of Next-Generation Sequencing in Pharmacogenetics and Pharmacogenomics. Cold Spring Harb. Perspect. Med. 2019, 9, a033027. [Google Scholar] [CrossRef] [PubMed]
- Dengu, F. Next-generation sequencing methods to detect donor-derived cell-free DNA after transplantation. Transplant. Rev. 2020, 34, 100542. [Google Scholar] [CrossRef]
- Zhao, Y.; Xia, Q.; Yin, Y.; Wang, Z. Comparison of Droplet Digital PCR and Quantitative PCR Assays for Quantitative Detection of Xanthomonas citri Subsp. citri. PLoS ONE 2016, 11, e0159004. [Google Scholar] [CrossRef] [PubMed]
- Feingold, B.; Rose-Felker, K.; West, S.C.; Zinn, M.D.; Berman, P.; Moninger, A.; Huston, A.; Stinner, B.; Xu, Q.; Zeevi, A.; et al. Early findings after integration of donor-derived cell-free DNA into clinical care following pediatric heart transplantation. Pediatr. Transplant. 2022, 26, e14124. [Google Scholar] [CrossRef] [PubMed]
- Amadio, J.M.; Rodenas-Alesina, E.; Superina, S.; Kozuszko, S.; Tsang, K.; Simard, A.; Aleksova, N.; Kobulnik, J.; Fan, C.-P.S.; Wijeysundera, H.C.; et al. Sparing the Prod: Providing an Alternative to Endomyocardial Biopsies with Noninvasive Surveillance After Heart Transplantation During COVID-19. CJC Open 2022, 4, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Kamath, M.; Shekhtman, G.; Grogan, T.; Hickey, M.J.; Silacheva, I.; Shah, K.S.; Shah, K.S.; Hairapetian, A.; Gonzalez, D.; Godoy, G.; et al. Variability in Donor-Derived Cell-Free DNA Scores to Predict Mortality in Heart Transplant Recipients—A Proof-of-Concept Study. Front. Immunol. 2022, 13, 825108. [Google Scholar] [CrossRef]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. BioTechniques 1992, 13, 444–449. [Google Scholar]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Ottesen, E.A.; Hong, J.W.; Quake, S.R.; Leadbetter, J.R. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 2006, 314, 1464–1467. [Google Scholar] [CrossRef]
- Morrison, T.; Hurley, J.; Garcia, J.; Yoder, K.; Katz, A.; Roberts, D.; Cho, J.; Kanigan, T.; Ilyin, S.E.; Horowitz, D.; et al. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006, 34, e123. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, S.O.; Wittwer, C.T.; Gao, C.; Gale, B.K. Spinning disk platform for microfluidic digital polymerase chain reaction. Anal. Chem. 2010, 82, 1546–1550. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, J.G.H.P.; Peeters, A.M.A.; Hesselink, D.A.; Boer, K. Pitfalls in the Detection of Donor-Derived Cell-Free DNA in Transplant Recipients. Clin. Chem. 2021, 67, 1030–1032. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, J.G.H.P.; Baan, C.C.; Peeters, A.M.A.; Nieboer, D.; Hesselink, D.A.; Boer, K. A comparison of two different analytical methods for donor-derived cell-free DNA quantification. Clin. Biochem. 2021, 96, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Oellerich, M.; Christenson, R.H.; Beck, J.; Schütz, E.; Sherwood, K.; Price, C.P.; Keown, P.A.; Walson, P.D. Donor-Derived Cell-Free DNA Testing in Solid Organ Transplantation: A Value Proposition. J. Appl. Lab. Med. 2020, 5, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Picard, C.; Frassati, C.; Cherouat, N.; Maioli, S.; Moskovtchenko, P.; Cherel, M.; Chiaroni, J.; Pedini, P. New methods for the quantification of mixed chimerism in transplantation. Front. Immunol. 2023, 14, 1023116. [Google Scholar] [CrossRef] [PubMed]
- Horgan, D.; Čufer, T.; Gatto, F.; Lugowska, I.; Verbanac, D.; Carvalho, Â.; Lal, J.A.; Kozaric, M.; Toomey, S.; Ivanov, H.Y.; et al. Accelerating the Development and Validation of Liquid Biopsy for Early Cancer Screening and Treatment Tailoring. Healthcare 2022, 10, 1714. [Google Scholar] [CrossRef]
- Lockwood, C.M.; Borsu, L.; Cankovic, M.; Earle, J.S.L.; Gocke, C.D.; Hameed, M.; Jordan, D.; Lopategui, J.R.; Pullambhatla, M.; Reuther, J.; et al. Recommendations for Cell-Free DNA Assay Validations: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2023, 25, 876–897. [Google Scholar] [CrossRef]
- Abedalthagafi, M.; Bawazeer, S.; Fawaz, R.I.; Heritage, A.M.; Alajaji, N.M.; Faqeih, E. Non-invasive prenatal testing: A revolutionary journey in prenatal testing. Front. Med. 2023, 10, 1265090. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorbini, M.; Carradori, T.; Togliatto, G.M.; Vaisitti, T.; Deaglio, S. Technical Advances in Circulating Cell-Free DNA Detection and Analysis for Personalized Medicine in Patients’ Care. Biomolecules 2024, 14, 498. https://doi.org/10.3390/biom14040498
Sorbini M, Carradori T, Togliatto GM, Vaisitti T, Deaglio S. Technical Advances in Circulating Cell-Free DNA Detection and Analysis for Personalized Medicine in Patients’ Care. Biomolecules. 2024; 14(4):498. https://doi.org/10.3390/biom14040498
Chicago/Turabian StyleSorbini, Monica, Tullia Carradori, Gabriele Maria Togliatto, Tiziana Vaisitti, and Silvia Deaglio. 2024. "Technical Advances in Circulating Cell-Free DNA Detection and Analysis for Personalized Medicine in Patients’ Care" Biomolecules 14, no. 4: 498. https://doi.org/10.3390/biom14040498
APA StyleSorbini, M., Carradori, T., Togliatto, G. M., Vaisitti, T., & Deaglio, S. (2024). Technical Advances in Circulating Cell-Free DNA Detection and Analysis for Personalized Medicine in Patients’ Care. Biomolecules, 14(4), 498. https://doi.org/10.3390/biom14040498






