Caspase-1 Deficiency Modulates Adipogenesis through Atg7-Mediated Autophagy: An Inflammatory-Independent Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. Glucose and Insulin Tolerance Tests
2.3. RNA Preparation and RT-qPCR
2.4. Western Blotting and Antibodies
2.5. Cell Culture and Induction of Differentiation
2.6. Isolation of SVF and SVF Culture (Primary Adipocytes)
2.7. RNA Interference
2.8. Oil Red O Staining and Quantification
2.9. Hematoxylin and Eosin (H&E) Staining
2.10. Statistical Analysis
3. Results
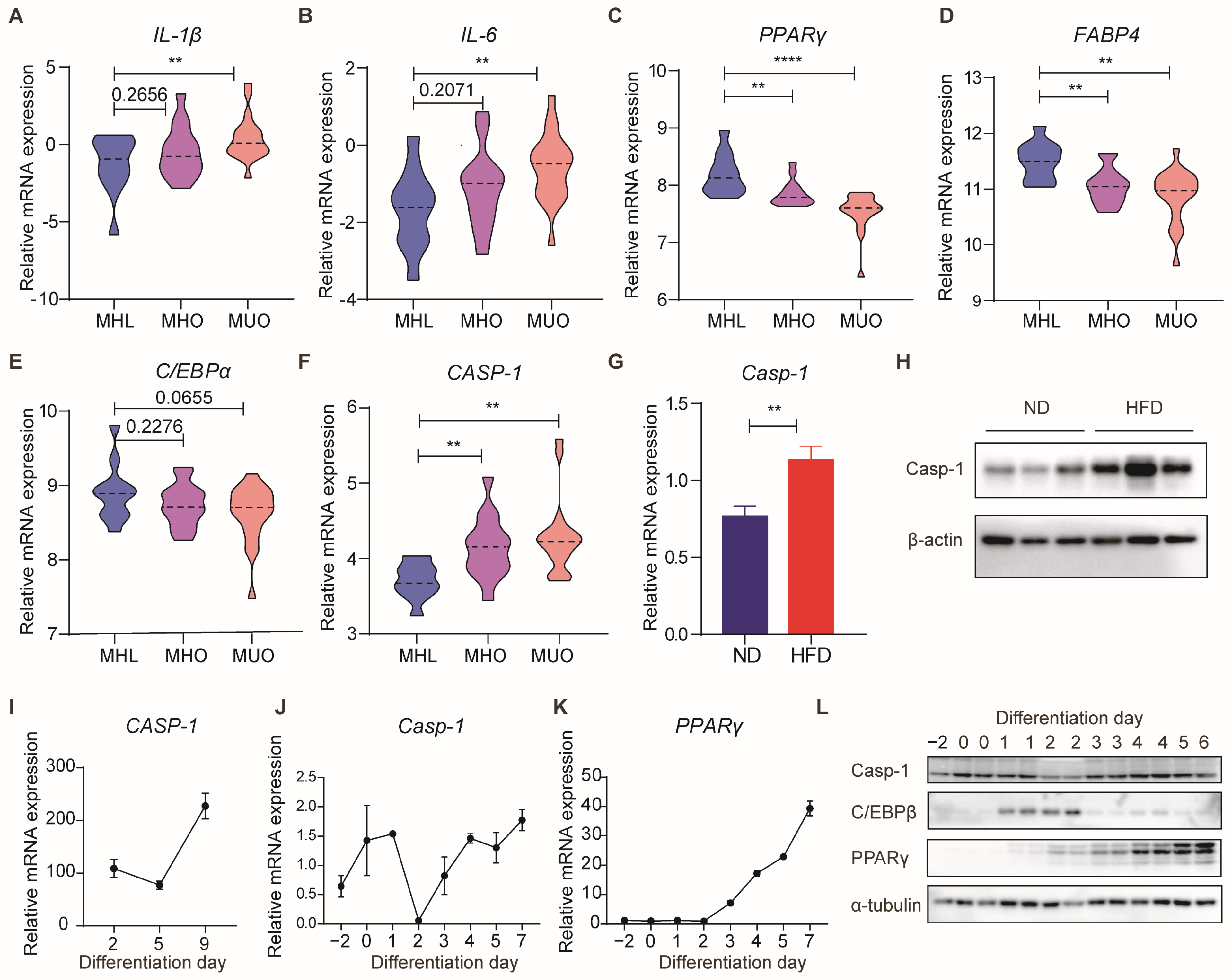
3.1. Caspase-1 Expression Is Correlated with Obesity and Adipogenesis
3.2. Deficiency of Caspase-1 Improves Glucose Homeostasis, Accompanied by an Increase in White Adipose Tissue, under a Normal Diet
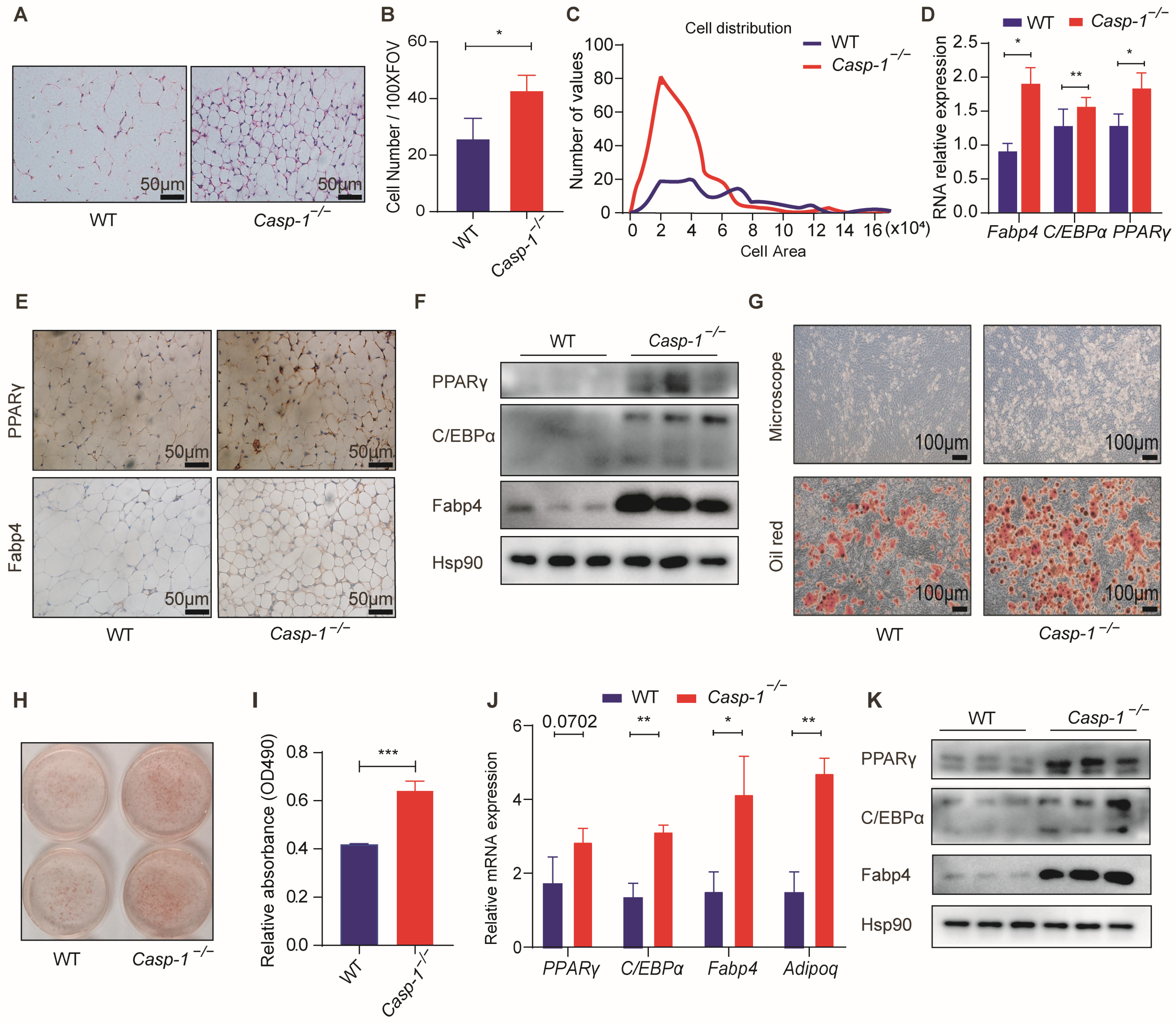
3.3. Caspase-1 Deficiency Promotes Adipogenesis In Vitro and In Vivo
3.4. Deficiency of Caspase-1 Enhances Autophagy in Adipocytes
3.5. Deficiency of Caspase-1 Promotes Adipogenesis by Enhancing Autophagy
3.6. Caspase-1 Deficiency Enhances Autophagy and Adipogenesis through Atg7
3.7. Caspase-1 Deficiency Ameliorates High-Fat Diet-Induced Obesity and Glucose Intolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boutari, C.; Mantzoros, C.S. A 2022 update on the epidemiology of obesity and a call to action: As its twin COVID-19 pandemic appears to be receding, the obesity and dysmetabolism pandemic continues to rage on. Metabolism 2022, 133, 155217. [Google Scholar] [CrossRef] [PubMed]
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.C.; Ding, C.; Magkos, F. The epidemiology of obesity. Metabolism 2019, 92, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Boutari, C.; Mantzoros, C.S. Inflammation: A key player linking obesity with malignancies. Metabolism 2018, 81, A3–A6. [Google Scholar] [CrossRef] [PubMed]
- Ruze, R.; Liu, T.; Zou, X.; Song, J.; Chen, Y.; Xu, R.; Yin, X.; Xu, Q. Obesity and type 2 diabetes mellitus: Connections in epidemiology, pathogenesis, and treatments. Front. Endocrinol. 2023, 14, 1161521. [Google Scholar] [CrossRef] [PubMed]
- Zechner, R.; Zimmermann, R.; Eichmann, T.O.; Kohlwein, S.D.; Haemmerle, G.; Lass, A.; Madeo, F. FAT SIGNALS—Lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012, 15, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From stem cell to adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef] [PubMed]
- Hammarstedt, A.; Gogg, S.; Hedjazifar, S.; Nerstedt, A.; Smith, U. Impaired Adipogenesis and Dysfunctional Adipose Tissue in Human Hypertrophic Obesity. Physiol. Rev. 2018, 98, 1911–1941. [Google Scholar] [CrossRef]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Investig. 2019, 129, 4022–4031. [Google Scholar] [CrossRef]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef]
- de Sá, P.M.; Richard, A.J.; Hang, H.; Stephens, J.M. Transcriptional Regulation of Adipogenesis. Compr. Physiol. 2011, 7, 635–674. [Google Scholar]
- Ganbold, M.; Ferdousi, F.; Arimura, T.; Tominaga, K.; Isoda, H. New Amphiphilic Squalene Derivative Improves Metabolism of Adipocytes Differentiated From Diabetic Adipose-Derived Stem Cells and Prevents Excessive Lipogenesis. Front. Cell Dev. Biol. 2020, 8, 577259. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.Z.; Althagafi, I.I.; Shamshad, H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur. J. Med. Chem. 2019, 166, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Sekar, M.; Thirumurugan, K. Autophagy: A molecular switch to regulate adipogenesis and lipolysis. Mol. Cell. Biochem. 2022, 477, 727–742. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Xiang, Y.; Wang, Y.; Baikati, K.; Cuervo, A.M.; Luu, Y.K.; Tang, Y.; Pessin, J.E.; Schwartz, G.J.; Czaja, M.J. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Investig. 2009, 119, 3329–3339. [Google Scholar] [CrossRef] [PubMed]
- Baerga, R.; Zhang, Y.; Chen, P.-H.; Goldman, S.; Jin, S.V. Targeted deletion ofautophagy-related 5(atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 2014, 5, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Engin, A.B. Adipocyte-Macrophage Cross-Talk in Obesity. Adv. Exp. Med. Biol. 2017, 960, 327–343. [Google Scholar] [CrossRef]
- Ye, J.; Gimble, J.M. Regulation of stem cell differentiation in adipose tissue by chronic inflammation. Clin. Exp. Pharmacol. Physiol. 2011, 38, 872–878. [Google Scholar] [CrossRef]
- Reilly, S.M.; Saltiel, A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017, 13, 633–643. [Google Scholar] [CrossRef]
- Jiang, N.; Li, Y.; Shu, T.; Wang, J. Cytokines and inflammation in adipogenesis: An updated review. Front. Med. 2019, 13, 314–329. [Google Scholar] [CrossRef]
- Herrera-Martínez, A.D.; Herrero-Aguayo, V.; Pérez-Gómez, J.M.; Gahete, M.D.; Luque, R.M. Inflammasomes: Cause or consequence of obesity-associated comorbidities in humans. Obesity 2022, 30, 2351–2362. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Karasawa, T.; Usui, F.; Kawashima, A.; Endo, Y.; Kobayashi, M.; Sadatomo, A.; Nakamura, J.; Iwasaki, Y.; Yada, T.; et al. Caspase-1 deficiency promotes high-fat diet-induced adipose tissue inflammation and the development of obesity. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E881–E890. [Google Scholar] [CrossRef]
- Wang, H.; Capell, W.; Yoon, J.H.; Faubel, S.; Eckel, R.H. Obesity development in caspase-1-deficient mice. Int. J. Obes. 2014, 38, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Stienstra, R.; Joosten, L.A.; Koenen, T.; van Tits, B.; van Diepen, J.A.; van den Berg, S.A.; Rensen, P.C.; Voshol, P.J.; Fantuzzi, G.; Hijmans, A.; et al. The inflammasome-mediated caspase-1 activation controls adipocyte differentiation and insulin sensitivity. Cell Metab. 2010, 12, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Autieri, M.V.; Scalia, R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol.-Cell Physiol. 2021, 320, C375–C391. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Olefsky, J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 2017, 127, 1–4. [Google Scholar] [CrossRef]
- Khan, F.; Khan, H.; Khan, A.; Yamasaki, M.; Moustaid-Moussa, N.; Al-Harrasi, A.; Rahman, S.M. Autophagy in adipogenesis: Molecular mechanisms and regulation by bioactive compounds. Biomed. Pharmacother. 2022, 155, 113715. [Google Scholar] [CrossRef]
- Ferhat, M.; Funai, K.; Boudina, S. Autophagy in Adipose Tissue Physiology and Pathophysiology. Antioxid. Redox Signal. 2019, 31, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Jabir, M.S.; Ritchie, N.D.; Li, D.; Bayes, H.K.; Tourlomousis, P.; Puleston, D.; Lupton, A.; Hopkins, L.; Simon, A.K.; Bryant, C.; et al. Caspase-1 Cleavage of the TLR Adaptor TRIF Inhibits Autophagy and β-Interferon Production during Pseudomonas aeruginosa Infection. Cell Host Microbe 2014, 15, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Shi, Y.; Tao, M.; Ma, X.; Hu, Y.; Huang, G.; Qiu, A.; Zhuang, S.; Liu, N. Delayed treatment with an autophagy inhibitor 3-MA alleviates the progression of hyperuricemic nephropathy. Cell Death Dis. 2020, 11, 467. [Google Scholar] [CrossRef]
- Pasquier, B. Autophagy inhibitors. Cell. Mol. Life Sci. 2015, 73, 985–1001. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; He, C. Pro-inflammatory cytokines: The link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 2018, 44, 38–50. [Google Scholar] [CrossRef]
- Xu, H.; Sethi, J.K.; Hotamisligil, G.S. Transmembrane Tumor Necrosis Factor (TNF)-α Inhibits Adipocyte Differentiation by Selectively Activating TNF Receptor 1. J. Biol. Chem. 1999, 274, 26287–26295. [Google Scholar] [CrossRef]
- Cawthorn, W.P.; Sethi, J.K. TNF-α and adipocyte biology. FEBS Lett. 2007, 582, 117–131. [Google Scholar] [CrossRef]
- Lagathu, C.; Bastard, J.-P.; Auclair, M.; Maachi, M.; Capeau, J.; Caron, M. Chronic interleukin-6 (IL-6) treatment increased IL-6 secretion and induced insulin resistance in adipocyte: Prevention by rosiglitazone. Biochem. Biophys. Res. Commun. 2003, 311, 372–379. [Google Scholar] [CrossRef]
- López, S. Interleukin-15 increases calcineurin expression in 3T3-L1 cells: Possible involvement on in vivo adipocyte differentiation. Int. J. Mol. Med. 2009, 24, 453–458. [Google Scholar] [CrossRef]
- Aguila, M.B.; Lacraz, G.; Rakotoarivelo, V.; Labbé, S.M.; Vernier, M.; Noll, C.; Mayhue, M.; Stankova, J.; Schwertani, A.; Grenier, G.; et al. Deficiency of Interleukin-15 Confers Resistance to Obesity by Diminishing Inflammation and Enhancing the Thermogenic Function of Adipose Tissues. PLoS ONE 2016, 11, e0162995. [Google Scholar] [CrossRef]
- Neal, J.W.; Clipstone, N.A. Calcineurin Mediates the Calcium-dependent Inhibition of Adipocyte Differentiation in 3T3-L1 Cells. J. Biol. Chem. 2002, 277, 49776–49781. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xu, W.; Zhou, R. NLRP3 inflammasome activation and cell death. Cell. Mol. Immunol. 2021, 18, 2114–2127. [Google Scholar] [CrossRef] [PubMed]
- Strowig, T.; Henao-Mejia, J.; Elinav, E.; Flavell, R. Inflammasomes in health and disease. Nature 2012, 481, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Simons, P.J.; van den Pangaart, P.S.; van Roomen, C.P.A.A.; Aerts, J.M.F.G.; Boon, L. Cytokine-mediated modulation of leptin and adiponectin secretion during in vitro adipogenesis: Evidence that tumor necrosis factor-α- and interleukin-1β-treated human preadipocytes are potent leptin producers. Cytokine 2005, 32, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Solt, L.A.; Madge, L.A.; Orange, J.S.; May, M.J. Interleukin-1-induced NF-kappaB activation is NEMO-dependent but does not require IKKbeta. J. Biol. Chem. 2007, 282, 8724–8733. [Google Scholar] [CrossRef] [PubMed]
- Tanti, J.-F.; Jager, J. Cellular mechanisms of insulin resistance: Role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Curr. Opin. Pharmacol. 2009, 9, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Wood, I.S.; Wang, B.; Jenkins, J.R.; Trayhurn, P. The pro-inflammatory cytokine IL-18 is expressed in human adipose tissue and strongly upregulated by TNFα in human adipocytes. Biochem. Biophys. Res. Commun. 2005, 337, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.; Lewis, E.; Jensen, D.R.; Voshol, P.J.; Kullberg, B.J.; Tack, C.J.; van Krieken, H.; Kim, S.H.; Stalenhoef, A.F.; et al. Deficiency of interleukin-18 in mice leads to hyperphagia, obesity and insulin resistance. Nat. Med. 2006, 12, 650–656. [Google Scholar] [CrossRef]
- Denes, A.; Lopez-Castejon, G.; Brough, D. Caspase-1: Is IL-1 just the tip of the ICEberg? Cell Death Dis. 2012, 3, e338. [Google Scholar] [CrossRef]
- Molla, M.D.; Ayelign, B.; Dessie, G.; Geto, Z.; Admasu, T.D. Caspase-1 as a regulatory molecule of lipid metabolism. Lipids Health Dis. 2020, 19, 34. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Yao, H.; Shah, S.Z.A.; Wu, W.; Wang, D.; Zhao, Y.; Wang, L.; Zhou, X.; Zhao, D.; Yang, L. The NLRP3-Caspase 1 Inflammasome Negatively Regulates Autophagy via TLR4-TRIF in Prion Peptide-Infected Microglia. Front. Aging Neurosci. 2018, 10, 116. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Gao, W.; Loughran, P.; Shapiro, R.; Fan, J.; Billiar, T.R.; Scott, M.J. Caspase 1 Activation Is Protective against Hepatocyte Cell Death by Up-regulating Beclin 1 Protein and Mitochondrial Autophagy in the Setting of Redox Stress. J. Biol. Chem. 2013, 288, 15947–15958. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Huang, J.X.; Liu, Y.; Li, X.; Zhou, S.R.; Qian, S.W.; Liu, Y.; Zhu, H.; Huang, H.Y.; Dang, Y.J.; et al. Transactivation of Atg4b by C/EBPbeta promotes autophagy to facilitate adipogenesis. Mol. Cell Biol. 2013, 33, 3180–3190. [Google Scholar] [CrossRef]
- Kharaziha, P.; Panaretakis, T. Dynamics of Atg5–Atg12–Atg16L1 Aggregation and Deaggregation. In Molecular Characterization of Autophagic Responses, Part A; Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2017; pp. 247–255. [Google Scholar]
- Zhanga, Y.; Goldmana, S.; Baergaa, R.; Zhaoa, Y.; Komatsub, M.; Jin, S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 19860–19865. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Chen, G.; Xu, M.; Cui, Y.; He, W.; Zeng, H.; Zeng, T.; Cheng, R.; Li, X. Caspase-1 Deficiency Modulates Adipogenesis through Atg7-Mediated Autophagy: An Inflammatory-Independent Mechanism. Biomolecules 2024, 14, 501. https://doi.org/10.3390/biom14040501
Wang Y, Chen G, Xu M, Cui Y, He W, Zeng H, Zeng T, Cheng R, Li X. Caspase-1 Deficiency Modulates Adipogenesis through Atg7-Mediated Autophagy: An Inflammatory-Independent Mechanism. Biomolecules. 2024; 14(4):501. https://doi.org/10.3390/biom14040501
Chicago/Turabian StyleWang, Yumeng, Gaojun Chen, Min Xu, Yewei Cui, Weijiong He, Hongxiang Zeng, Ting Zeng, Rui Cheng, and Xi Li. 2024. "Caspase-1 Deficiency Modulates Adipogenesis through Atg7-Mediated Autophagy: An Inflammatory-Independent Mechanism" Biomolecules 14, no. 4: 501. https://doi.org/10.3390/biom14040501




