A Practical Guide for the Quality Evaluation of Fluobodies/Chromobodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construct Cloning
2.2. Protein Purification and Characterization
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Griep, R.A.; van Twisk, C.; van der Wolf, J.M.; Schots, A. Fluobodies: Green fluorescent single-chain Fv fusion proteins. J. Immunol. Methods 1999, 230, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Traenkle, B.; Rothbauer, U. Under the Microscope: Single-Domain Antibodies for Live-Cell Imaging and Super-Resolution Microscopy. Front. Immunol. 2017, 8, 1030. [Google Scholar] [CrossRef] [PubMed]
- Mazzega, E.; Beran, A.; Cabrini, M.; de Marco, A. In vitro isolation of nanobodies for selective Alexandrium minutum recognition: A model for convenient development of dedicated immuno-reagents to study and diagnostic toxic unicellular algae. Harmful Algae 2019, 82, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Oliinyk, O.S.; Baloban, M.; Clark, C.L.; Carey, E.; Pletnev, S.; Nimmerjahn, A.; Verkhusha, V.V. Single-domain near-infrared protein provides a scaffold for antigen-dependent fluorescent nanobodies. Nat. Methods 2022, 19, 740–750. [Google Scholar] [CrossRef]
- de Marco, A. Recombinant expression of nanobodies and nanobody-derived immunoreagents. Protein Expr. Purif. 2020, 172, 105645. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; de Marco, A. Improved quantitative and qualitative production of single-domain intrabodies mediated by the co-expression of Erv1p sulfhydryl oxidase. Protein Expr. Purif. 2011, 79, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Shemiakina, L.I.; Ermakova, G.V.; Cranfill, P.J.; Baird, M.A.; Evans, R.A.; Souslova, E.A.; Staroverov, D.B.; Gorokhovatsky, A.Y.; Putintseva, E.V.; Gorodnicheva, T.V.; et al. A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 2012, 3, 1204. [Google Scholar] [CrossRef]
- Pennacchietti, F.; Serebrovskaya, E.O.; Faro, A.R.; Shemyakina, I.I.; Bozhanova, N.G.; Kotlobay, A.A.; Gurskaya, N.G.; Bodén, A.; Dreier, J.; Chudakov, D.M.; et al. Fast reversibly photoswitching red fluorescent proteins for live-cell RESOLFT nanoscopy. Nat. Methods 2018, 15, 601–604. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, W.; Tan, L.; Chen, T.; He, Y.; Irving, P.S.; Weeks, K.M.; Zhang, Q.C.; Dong, X. Pervasive downstream RNA hairpins dynamically dictate start-codon selection. Nature 2023, 621, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Medenbach, J.; Seiler, M.; Hentze, M.W. Translational control via protein-regulated upstream open reading frames. Cell 2011, 145, 902–913. [Google Scholar] [CrossRef]
- Korandla, D.R.; Wozniak, J.M.; Campeau, A.; Gonzalez, D.J.; Wright, E.S. AssessORF: Combining evolutionary conservation and proteomics to assess prokaryotic gene predictions. Bioinformatics 2020, 36, 1022–1029. [Google Scholar] [CrossRef] [PubMed]
- Dimonaco, N.J.; Clare, A.; Kenobi, K.; Aubrey, W.; Creevey, C.J. StORF-Reporter: Finding genes between genes. Nucleic Acids Res. 2023, 51, 11504–11517. [Google Scholar] [CrossRef] [PubMed]
- Belinky, F.; Rogozin, I.B.; Koonin, E.V. Selection on start codons in prokaryotes and potential compensatory nucleotide substitutions. Sci. Rep. 2017, 7, 12422. [Google Scholar] [CrossRef]
- Chengguang, H.; Sabatini, P.; Brandi, L.; Giuliodori, A.M.; Pon, C.L.; Gualerzi, C.O. Ribosomal selection of mRNAs with degenerate initiation triplets. Nucleic Acids Res. 2017, 5, 7309–7325. [Google Scholar] [CrossRef] [PubMed]
- Muslinkina, L.; Pletnev, V.Z.; Pletneva, N.V.; Ruchkin, D.A.; Kolesov, D.V.; Bogdanov, A.M.; Kost, L.A.; Rakitina, T.V.; Agapova, Y.K.; Shemyakina, I.I.; et al. Two independent routes of post-translational chemistry in fluorescent protein FusionRed. Int. J. Biol. Macromol. 2020, 155, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Ando, R.; Shimozono, S.; Sugiyama, M.; Takeda, N.; Kurokawa, H.; Deguchi, R.; Endo, K.; Haga, K.; Takai-Todaka, R.; et al. A highly photostable and bright green fluorescent protein. Nat. Biotechnol. 2022, 40, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Ivorra-Molla, E.; Akhuli, D.; McAndrew, M.B.L.; Scott, W.; Kumar, L.; Palani, S.; Mishima, M.; Crow, A.; Balasubramanian, M.K. A monomeric StayGold fluorescent protein. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Ando, R.; Shimozono, S.; Ago, H.; Takagi, M.; Sugiyama, M.; Kurokawa, H.; Hirano, M.; Niino, Y.; Ueno, G.; Ishidate, F.; et al. StayGold variants for molecular fusion and membrane-targeting applications. Nat. Methods 2023, 21, 648–656. [Google Scholar] [CrossRef]
- Zhang, H.; Lesnov, G.D.; Subach, O.M.; Zhang, W.; Kuzmicheva, T.P.; Vlaskina, A.V.; Samygina, V.R.; Chen, L.; Ye, X.; Nikolaeva, A.Y.; et al. Bright and stable monomeric green fluorescent protein derived from StayGold. Nat. Methods 2024, 21, 657–665. [Google Scholar] [CrossRef]
- Djender, S.; Schneider, A.; Beugnet, A.; Crepin, R.; Desrumeaux, K.E.; Romani, C.; Moutel, S.; Perez, F.; de Marco, A. Bacterial cytoplasm as an effective cell compartment for producing functional VHH-based affinity reagents and Camelidae IgG-like recombinant antibodies. Microb. Cell Fact. 2014, 13, 140. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Snaith, H.A.; Anders, A.; Samejima, I.; Sawin, K.E. New and old reagents for fluorescent protein tagging of microtubules in fission yeast; experimental and critical evaluation. Methods Cell Biol. 2010, 97, 147–172. [Google Scholar] [PubMed]
- Campbell, B.C.; Paez-Segala, M.G.; Looger, L.L.; Petsko, G.A.; Liu, C.F. Chemically stable fluorescent proteins for advanced microscopy. Nat. Methods 2022, 19, 1612–1621. [Google Scholar] [CrossRef] [PubMed]
- Cranfill, P.J.; Sell, B.R.; Baird, M.A.; Allen, J.R.; Lavagnino, Z.; de Gruiter, H.M.; Kremers, G.J.; Davidson, M.W.; Ustione, A.; Piston, D.W. Quantitative assessment of fluorescent proteins. Nat. Methods 2016, 13, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Martynov, V.I.; Savitsky, A.P.; Martynova, N.Y.; Savitsky, P.A.; Lukyanov, K.A.; Lukyanov, S.A. Alternative cyclization in GFP-like proteins family. The formation and structure of the chromophore of a purple chromoprotein from Anemonia sulcata. J. Biol. Chem. 2001, 276, 21012–21016. [Google Scholar] [CrossRef]
- Zagranichny, V.E.; Rudenko, N.V.; Gorokhovatsky, A.Y.; Zakharov, M.V.; Balashova, T.A.; Arseniev, A.S. Traditional GFP-type cyclization and unexpected fragmentation site in a purple chromoprotein from Anemonia sulcata, asFP595. Biochemistry 2004, 43, 13598–13603. [Google Scholar] [CrossRef] [PubMed]
- Aronson, D.E.; Costantini, L.M.; Snapp, E.L. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic 2011, 12, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Meiresonne, N.Y.; Consoli, E.; Mertens, L.M.Y.; Chertkova, A.O.; Goedhart, J.; den Blaauwen, T. Superfolder mTurquoise2ox optimized for the bacterial periplasm allows high efficiency in vivo FRET of cell division antibiotic targets. Mol. Microbiol. 2019, 111, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; Giabbai, B.; Semrau, M.S.; Medagli, B.; Riccio, V.; Bajc, G.; Storici, P.; de Marco, A. Comparative analysis of fusion tags used to functionalize recombinant antibodies. Protein Expr. Purif. 2020, 166, 105505. [Google Scholar] [CrossRef]
- Shimozono, S.; Ando, R.; Sugiyama, M.; Hirano, M.; Niino, Y.; Miyawaki, A. Comparison of monomeric variants of StayGold. bioRxiv 2024. [Google Scholar] [CrossRef]
- D’Ercole, C.; De March, M.; Veggiani, G.; Oloketuyi, S.; Svigelj, R.; de Marco, A. Biological applications of synthetic binders isolated from a conceptually new adhiron library. Biomolecules 2023, 13, 1533. [Google Scholar] [CrossRef] [PubMed]
- Matlashov, M.E.; Shcherbakova, D.M.; Alvelid, J.; Baloban, M.; Pennacchietti, F.; Shemetov, A.A.; Testa, I.; Verkhusha, V.V. A set of monomeric near-infrared fluorescent proteins for multicolor imaging across scales. Nat. Commun. 2020, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Grishin, N.V. PROMALS3D: Multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 2014, 1079, 263–271. [Google Scholar] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]

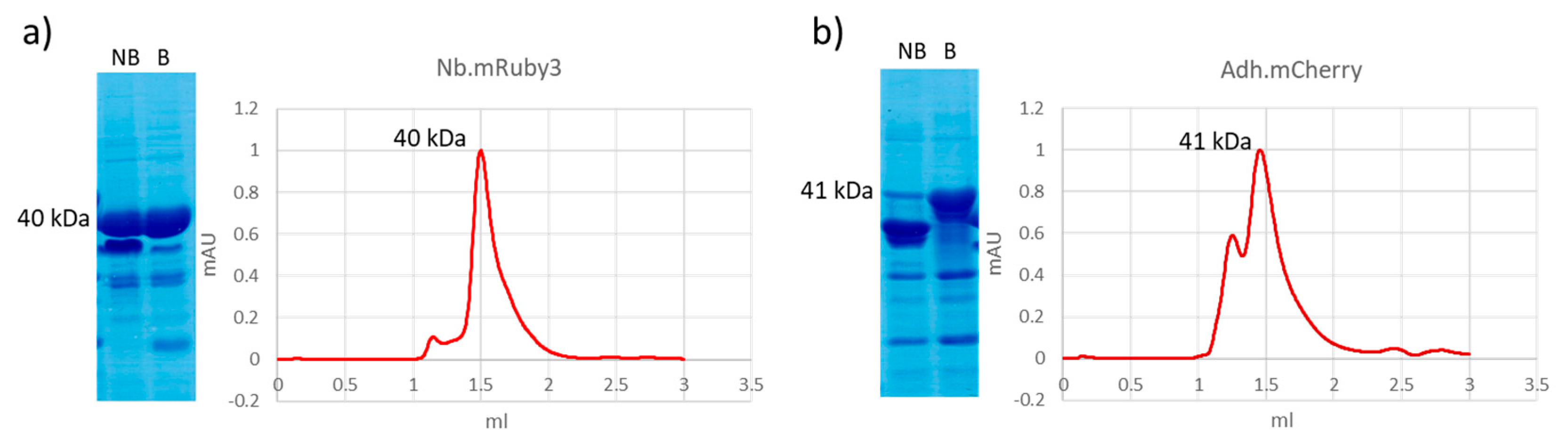
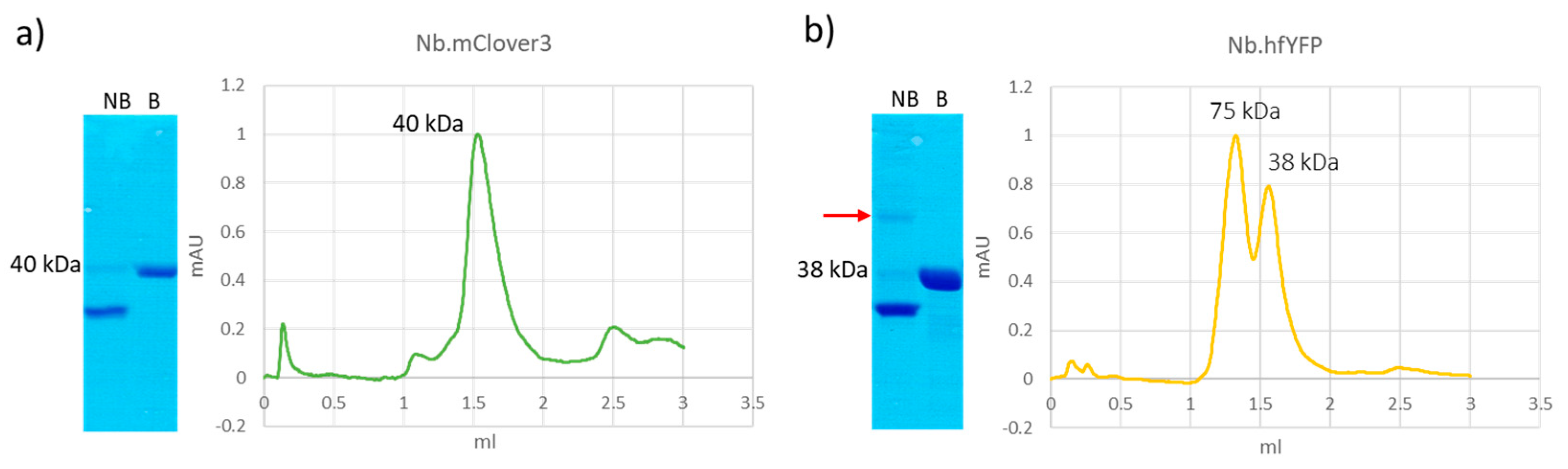

| Original Fluorescent Protein | Derived Proteins | Degradation Products in SDS-PAGE |
|---|---|---|
| eqFP611 (Entacmaea quadricolor) | mRuby3, Electra | Yes |
| avGFP (Aequorea victoria) | mClover3, hfYFP, mEGFP | No |
| dsRed (Discosoma spp.) | mCherry, tdTomato, mBlueberry2 | Yes |
| CU17s (Cytaeis uchidae) | StayGold, monomeric StayGold variants | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Štrancar, U.; D’Ercole, C.; Cikatricisová, L.; Nakić, M.; De March, M.; de Marco, A. A Practical Guide for the Quality Evaluation of Fluobodies/Chromobodies. Biomolecules 2024, 14, 587. https://doi.org/10.3390/biom14050587
Štrancar U, D’Ercole C, Cikatricisová L, Nakić M, De March M, de Marco A. A Practical Guide for the Quality Evaluation of Fluobodies/Chromobodies. Biomolecules. 2024; 14(5):587. https://doi.org/10.3390/biom14050587
Chicago/Turabian StyleŠtrancar, Urša, Claudia D’Ercole, Lucia Cikatricisová, Mirna Nakić, Matteo De March, and Ario de Marco. 2024. "A Practical Guide for the Quality Evaluation of Fluobodies/Chromobodies" Biomolecules 14, no. 5: 587. https://doi.org/10.3390/biom14050587







