SUMOylation in Giardia lamblia: A Conserved Post-Translational Modification in One of the Earliest Divergent Eukaryotes
Abstract
:1. Introduction
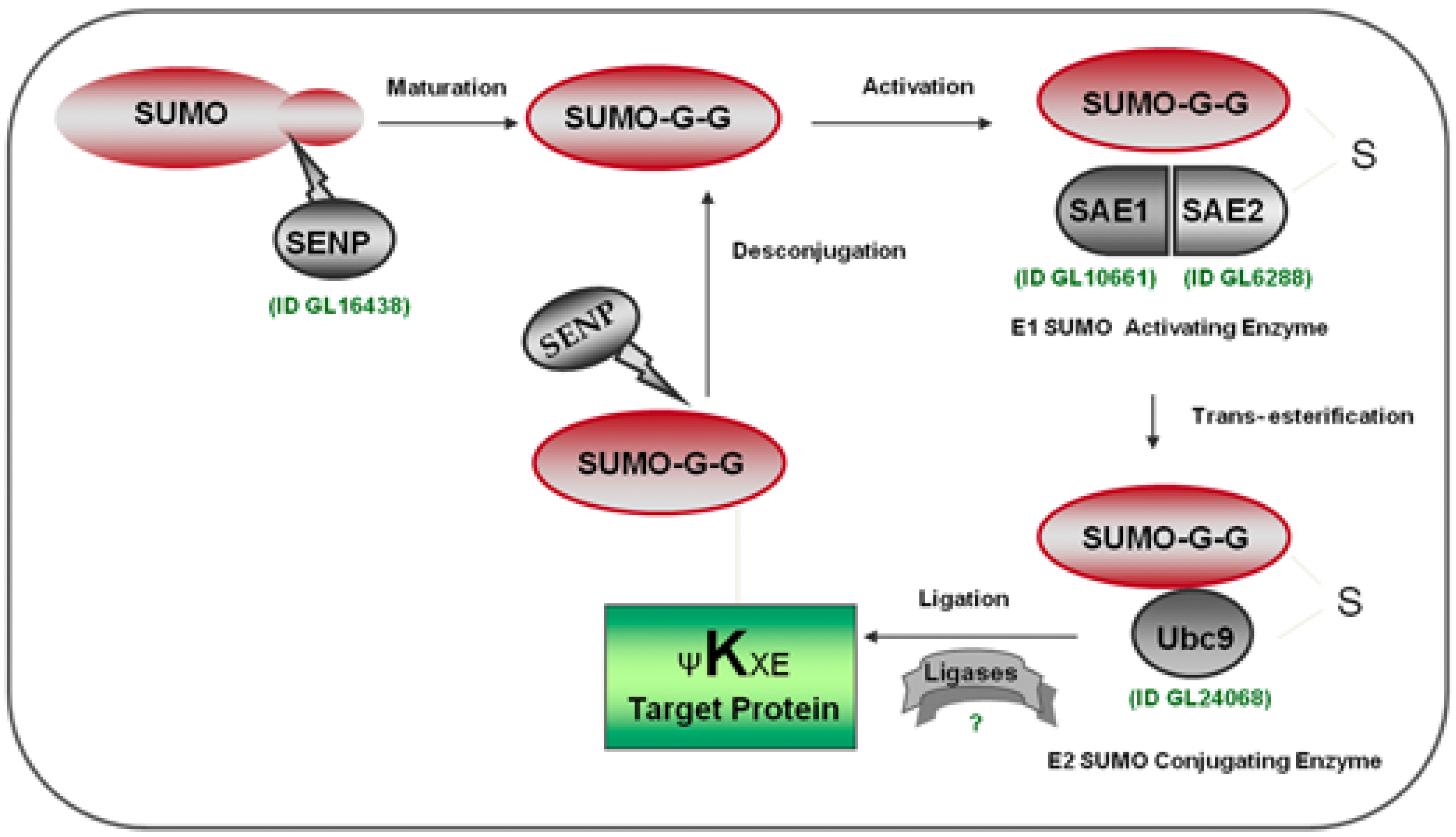
2. Results and Discussion
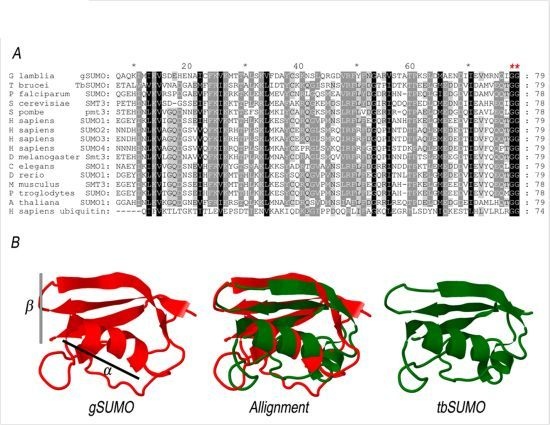
2.1. Characterization of SUMO in Giardia lamblia
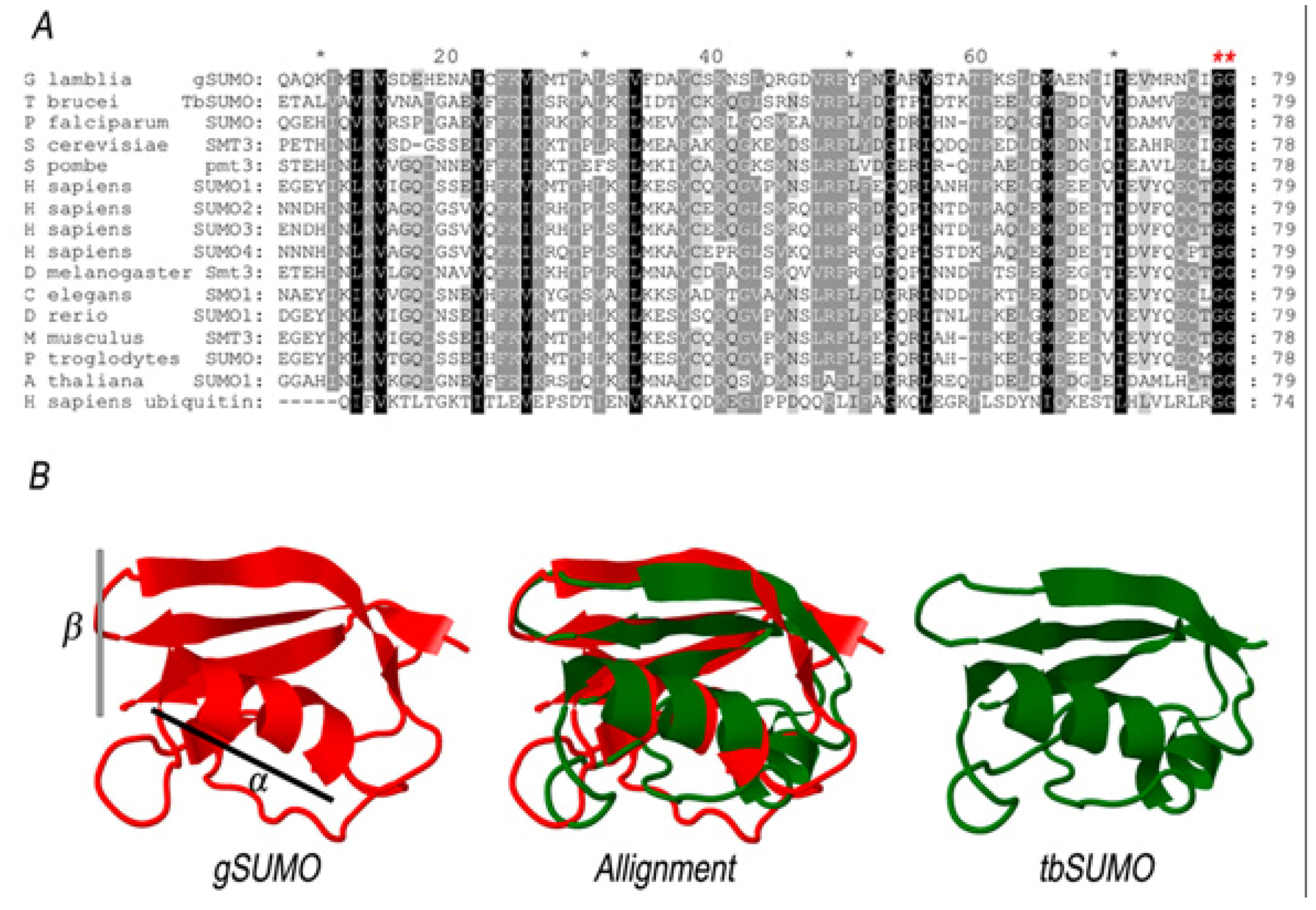

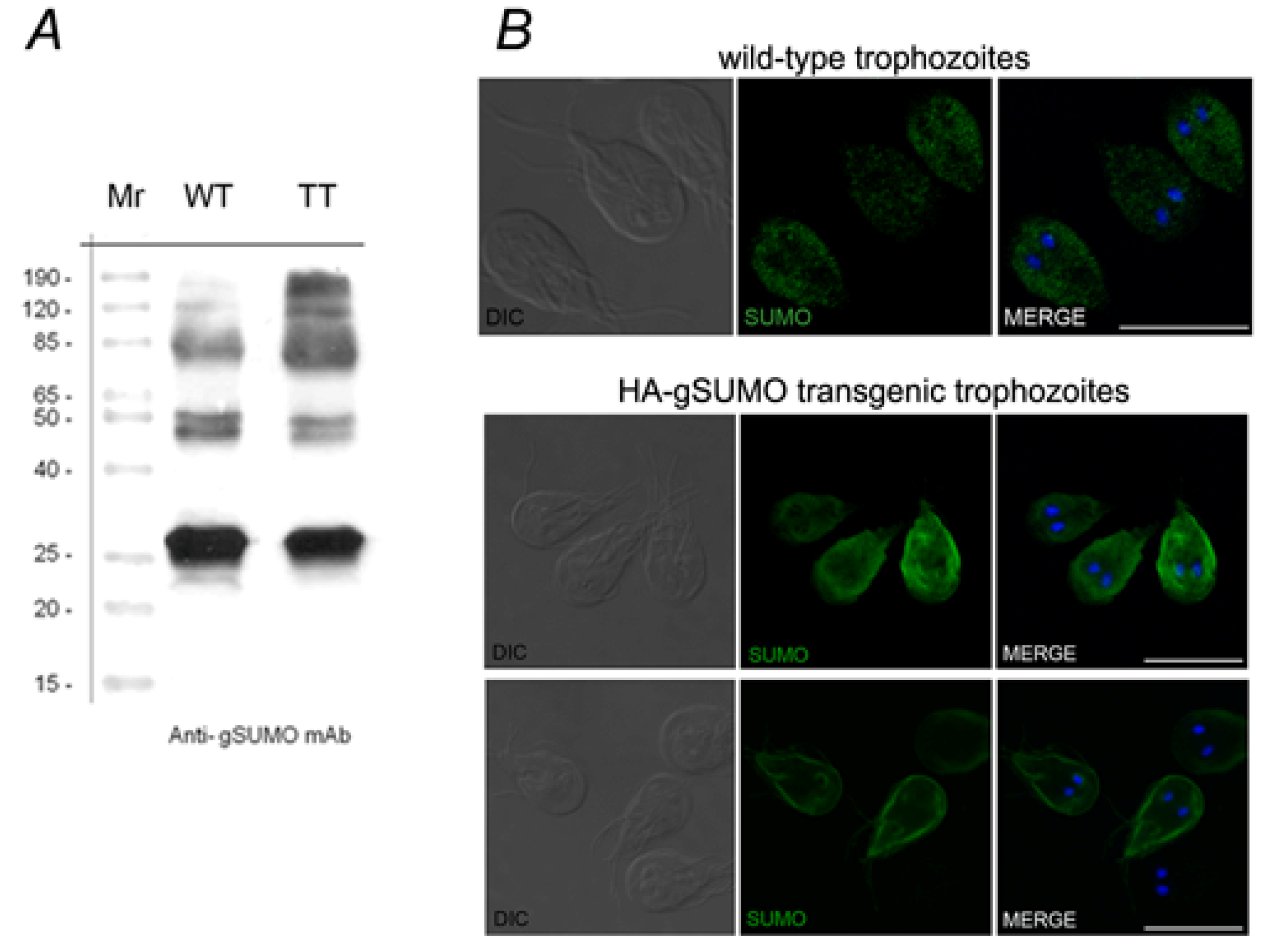
2.2. Identification of the SUMOylation Pathway Components
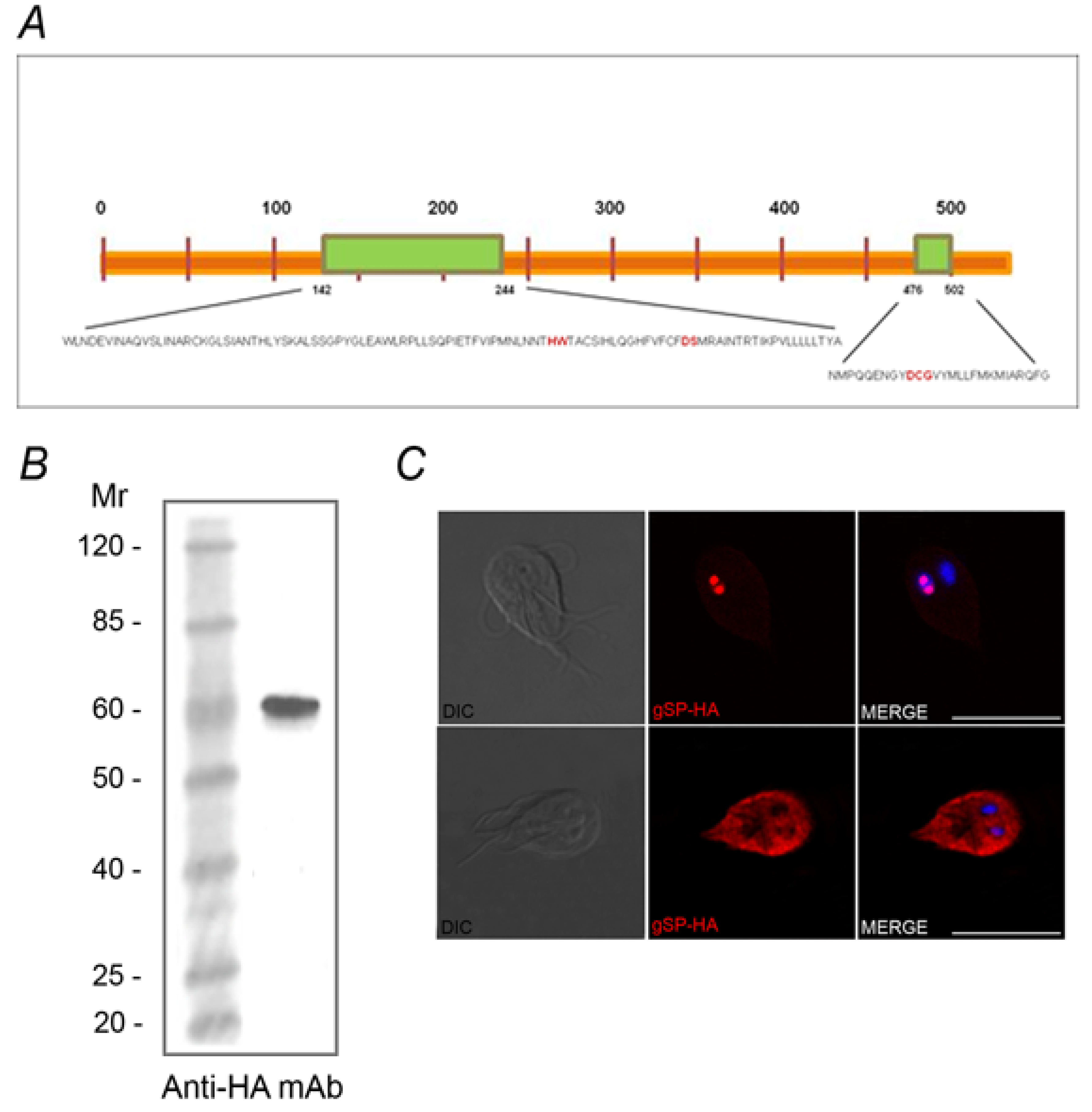
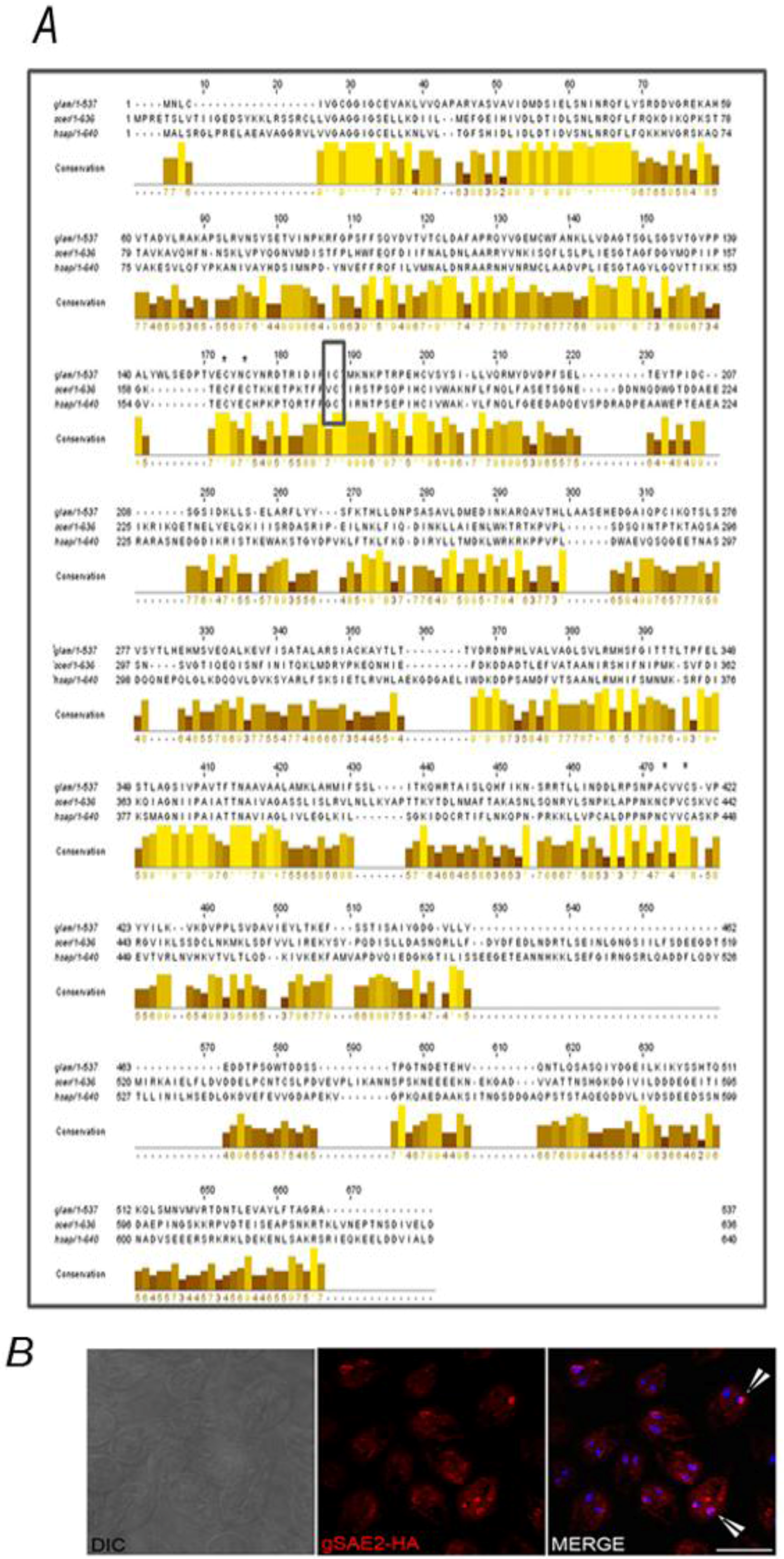
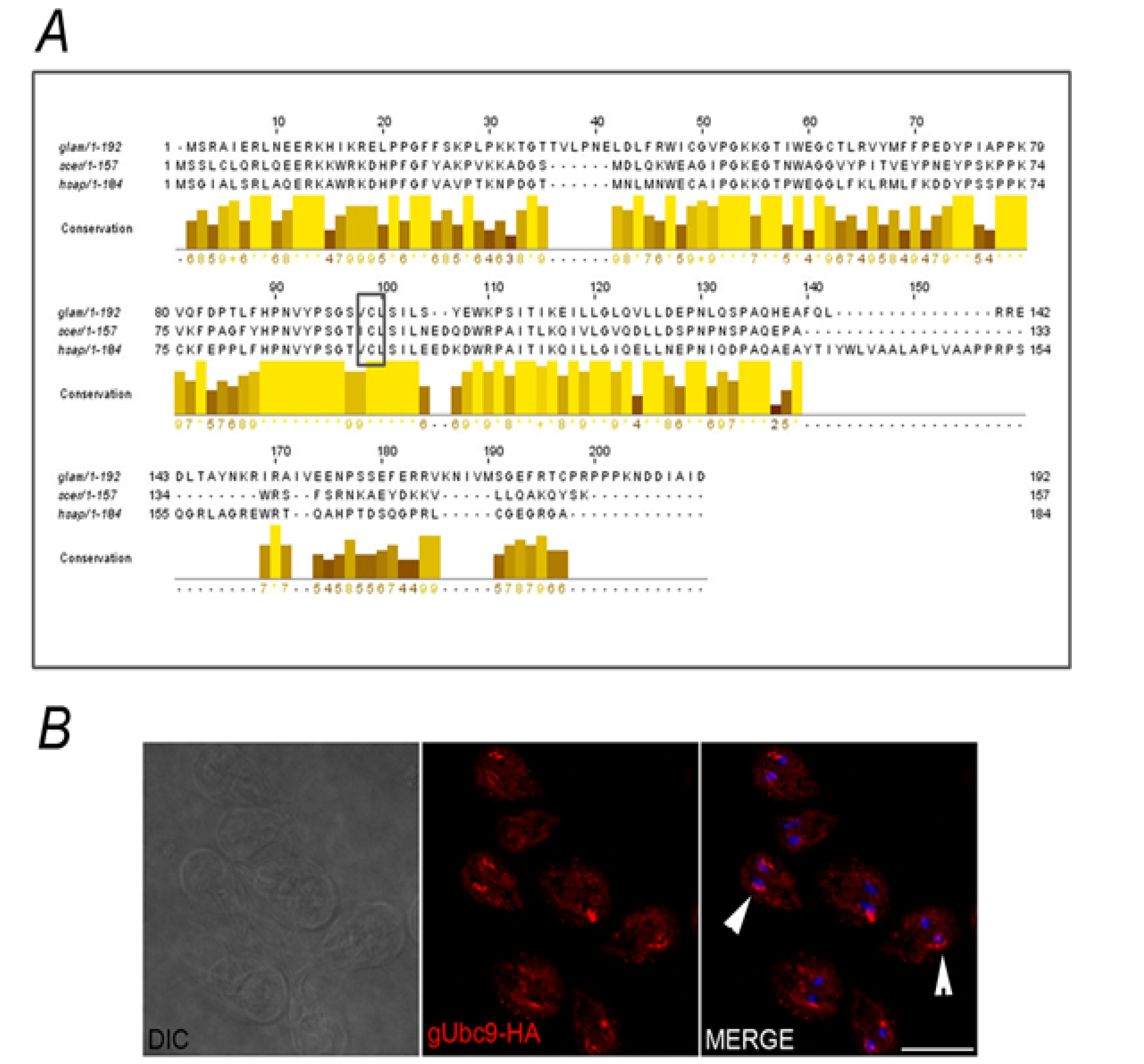
3. Experimental Section
3.1. Parasites, Cells and Media
3.2. Mice
3.3. Bioinformatics Analysis
3.4. Isolation of gsumo; gsp; gsae2; gubc9
3.5. Plasmid Construction by gSUMO Over-expression
3.6. Recombinant Protein gSUMO Fused to GST Expression and Purification
3.7. Monoclonal Antibody Production
3.8. Western Blotting
3.9. Immunofluorescence Assay
4. Conclusions
Acknowledgments
Author Contribution
Conflict of Insterest
References
- Farthing, M.J. The molecular pathogenesis of giardiasis. J. Pediatr. Gastroenterol. Nutr. 1997, 24, 79–88. [Google Scholar] [CrossRef]
- dam, R.D. Biology of Giardia lamblia. Clin. Microbiol. Rev. 2001, 14, 447–475. [Google Scholar] [CrossRef]
- Lujan, H.D. Mechanisms of adaptation in the intestinal parasite Giardia lamblia. Essays Biochem. 2011, 51, 177–191. [Google Scholar]
- Morrison, H.G.; McArthur, A.G.; Gillin, F.D.; Aley, S.B.; Adam, R.D.; Olsen, G.J.; Best, A.A.; Cande, W.Z.; Chen, F.; Cipriano, M.J.; et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Sci. 2007, 317, 1921–1926. [Google Scholar]
- Muller, S.; Hoege, C.; Pyrowolakis, G.; Jentsch, S. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2001, 2, 202–210. [Google Scholar] [CrossRef]
- Schwartz, D.C.; Hochstrasser, M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 2003, 28, 321–328. [Google Scholar] [CrossRef]
- Leach, M.D.; Stead, D.A.; Argo, E.; Brown, A.J. Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology, and stress resistance in the pathogen Candida albicans. Mol. Biol. Cell 2011, 22, 687–702. [Google Scholar] [CrossRef]
- Gill, G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004, 18, 2046–2059. [Google Scholar] [CrossRef]
- Melchior, F. SUMO--nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000, 16, 591–626. [Google Scholar] [CrossRef]
- Dorval, V.; Fraser, P.E. SUMO on the road to neurodegeneration. Biochim. Biophys. Acta 2007, 1773, 694–706. [Google Scholar] [CrossRef]
- Bossis, G.; Melchior, F. SUMO: regulating the regulator. Cell Div. 2006, 1–13. [Google Scholar]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef]
- Moutty, M.C.; Sakin, V.; Melchior, F. Importin alpha/beta mediates nuclear import of individual SUMO E1 subunits and of the holo-enzyme. Mol. Biol. Cell. 2011, 22, 652–660. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Y. Role of the Zn(2+) motif of E1 in SUMO adenylation. J. Biol. Chem. 2010, 285, 23732–23738. [Google Scholar] [CrossRef]
- Okada, S.; Nagabuchi, M.; Takamura, Y.; Nakagawa, T.; Shinmyozu, K.; Nakayama, J.; Tanaka, K. Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 2009, 50, 1049–1061. [Google Scholar] [CrossRef]
- Lois, L.M.; Lima, C.D. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005, 24, 439–451. [Google Scholar] [CrossRef]
- Tatham, M.H.; Chen, Y.; Hay, R.T. Role of two residues proximal to the active site of Ubc9 in substrate recognition by the Ubc9.SUMO-1 thiolester complex. Biochemistry 2003, 42, 3168–3179. [Google Scholar]
- Seeler, J.S.; Dejean, A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 2003, 4, 690–699. [Google Scholar] [CrossRef]
- Braun, L.; Cannella, D.; Pinheiro, A.M.; Kieffer, S.; Belrhali, H.; Garin, J.; Hakini, M.A. The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int. J. Parasitol. 2009, 39, 81–90. [Google Scholar] [CrossRef]
- Issar, N.; Roux, E.; Mattei, D.; Scherf, A. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell Microbiol. 2008, 10, 1999–2011. [Google Scholar] [CrossRef]
- Bayona, J.C.; Nakayasu, E.S.; Laverriere, M.; Aguilar, C.; Sobreira, T.J.; Choi, H.; Nesvizhsdii, A.I.; Almeida, I.C.; Cazzulo, J.J.; Alvarez, V.E. SUMOylation pathway in Trypanosoma cruzi: functional characterization and proteomic analysis of target proteins. Mol. Cell Proteomics 2011, 10, M110–007369. [Google Scholar]
- Shang, Q.; Xu, C.; Zhang, J.; Zhang, X.; Tu, X. Solution structure of SUMO from Trypanosoma brucei and its interaction with Ubc9. Proteins 2009, 76, 266–269. [Google Scholar] [CrossRef]
- Touz, M.C.; Ropolo, A.S.; Rivero, M.R.; Vranych, C.V.; Conrad, J.T.; Svard, S.G.; Nash, T.E. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 2008, 121, 2930–2938. [Google Scholar] [CrossRef]
- Rosonina, E.; Duncan, S.M.; Manley, J.L. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010, 24, 1242–1252. [Google Scholar] [CrossRef]
- Wang, Y.; Ladunga, I.; Miller, A.R.; Horken, K.M.; Plucinak, T.; Weeks, D.P.; Bailey, C.P. The small ubiquitin-like modifier (SUMO) and SUMO-conjugating system of Chlamydomonas reinhardtii. Genetics 2008, 179, 177–192. [Google Scholar]
- Liao, S.; Wang, T.; Fan, K.; Tu, X. The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp. Cell Res. 2010, 316, 704–715. [Google Scholar] [CrossRef]
- Kurepa, J.; Walker, J.M.; Smalle, J.; Gosink, M.M.; Davis, S.J.; Durham, T.L.; Sung, D.Y.; Vierstra, R.D. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem 2003, 278, 6862–6872. [Google Scholar]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef]
- Colby, T.; Matthai, A.; Boeckelmann, A.; Stuible, H.P. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006, 142, 318–332. [Google Scholar]
- Li, T.; Evdokimov, E.; Shen, R.F.; Chao, C.C.; Tekle, E.; Wang, T.; Stadtman, E.R.; Yang, D.C.; Chorck, P.B. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad. Sci. USA 2004, 101, 8551–8556. [Google Scholar]
- Ponder, E.L.; Albrow, V.E.; Leader, B.A.; Bekes, M.; Mikolajczyk, J.; Fonovic, U.P.; Shen, A.; Drag, M.; Xiao, J.; Deu, E.; et al. Functional characterization of a SUMO deconjugating protease of Plasmodium falciparum using newly identified small molecule inhibitors. Chem. Biol. 2011, 18, 711–721. [Google Scholar]
- Wegscheid-Gerlach, C.; Gerber, H.D.; Diederich, W.E. Proteases of Plasmodium falciparum as potential drug targets and inhibitors thereof. Curr. Top Med. Chem. 2010, 10, 346–367. [Google Scholar] [CrossRef]
- Bailey, D.; O'Hare, P. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 2002, 83, 2951–2964. [Google Scholar]
- Hang, J.; Dasso, M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 2002, 277, 19961–19966. [Google Scholar] [CrossRef]
- Zhang, H.; Saitoh, H.; Matunis, M.J. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell Biol. 2002, 22, 6498–6508. [Google Scholar] [CrossRef]
- Di Bacco, A.; Ouyang, J.; Lee, H.Y.; Catic, A.; Ploegh, H.; Gill, G. The SUMO-specific protease SENP5 is required for cell division. Mol. Cell Biol. 2006, 26, 4489–4498. [Google Scholar] [CrossRef]
- Wang, Y.; Dasso, M. SUMOylation and deSUMOylation at a glance. J. Cell Sci. 2009, 122, 4249–4252. [Google Scholar] [CrossRef]
- Johnson, E.S.; Schwienhorst, I.; Dohmen, R.J.; Blobel, G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997, 16, 5509–5519. [Google Scholar]
- Truong, K.; Lee, T.; Chen, Y. SUMO modification of the E1 Cys domain inhibits its enzymatic activity. J. Biol. Chem. 2012, 287, 15154–15163. [Google Scholar]
- Duan, X.; Trent, J.O.; Ye, H. Targeting the SUMO E2 conjugating enzyme Ubc9 interaction for anti-cancer drug design. Anticancer Agents Med Chem. 2009, 9, 51–54. [Google Scholar]
- Dasso, M. Emerging roles of the SUMO pathway in mitosis. Cell Div 2008, 3, 5. [Google Scholar] [CrossRef]
- Itahana, Y.; Yeh, E.T.; Zhang, Y. Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol. Cell Biol. 2006, 26, 4675–4689. [Google Scholar] [CrossRef]
- Keister, D.B. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983, 77, 487–488. [Google Scholar] [CrossRef]
- Singer, S.M.; Yee, J.; Nash, T.E. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol. Biochem Parasitol. 1998, 92, 59–69. [Google Scholar] [CrossRef]
- Yee, J.; Nash, T.E. Transient transfection and expression of firefly luciferase in Giardia lamblia. Proc. Natl. Acad. Sci. USA 1995, 92, 5615–5619. [Google Scholar]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar]
- Piast, M.; Kustrzeba-Wojcicka, I.; Matusiewicz, M.; Banas, T. Molecular evolution of enolase. Acta Biochim. Pol. 2005, 52, 507–513. [Google Scholar]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar]
- Abyzov, A.; Errami, M.; Leslin, C.M.; Ilyin, V.A. Friend, an integrated analytical front-end application for bioinformatics. Bioinformatics 2005, 21, 3677–3678. [Google Scholar]
- Sehr, P.; Zumbach, K.; Pawlita, M. A generic capture ELISA for recombinant proteins fused to glutathione S-transferase: validation for HPV serology. J. Immunol. Methods. 2001, 253, 153–162. [Google Scholar] [CrossRef]
- Gallego, E.; Alvarado, M.; Wasserman, M. Identification and expression of the protein ubiquitination system in Giardia intestinalis. Parasitol. Res. 2007, 101, 1–7. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vranych, C.V.; Merino, M.C.; Zamponi, N.; Touz, M.C.; Rópolo, A.S. SUMOylation in Giardia lamblia: A Conserved Post-Translational Modification in One of the Earliest Divergent Eukaryotes. Biomolecules 2012, 2, 312-330. https://doi.org/10.3390/biom2030312
Vranych CV, Merino MC, Zamponi N, Touz MC, Rópolo AS. SUMOylation in Giardia lamblia: A Conserved Post-Translational Modification in One of the Earliest Divergent Eukaryotes. Biomolecules. 2012; 2(3):312-330. https://doi.org/10.3390/biom2030312
Chicago/Turabian StyleVranych, Cecilia V., María C. Merino, Nahuel Zamponi, María C. Touz, and Andrea S. Rópolo. 2012. "SUMOylation in Giardia lamblia: A Conserved Post-Translational Modification in One of the Earliest Divergent Eukaryotes" Biomolecules 2, no. 3: 312-330. https://doi.org/10.3390/biom2030312
APA StyleVranych, C. V., Merino, M. C., Zamponi, N., Touz, M. C., & Rópolo, A. S. (2012). SUMOylation in Giardia lamblia: A Conserved Post-Translational Modification in One of the Earliest Divergent Eukaryotes. Biomolecules, 2(3), 312-330. https://doi.org/10.3390/biom2030312