Promiscuity of the Euonymus Carbohydrate-Binding Domain
Abstract
:1. Introduction
2. Lectins with an EUL Domain
2.1. Molecular Cloning of EEA
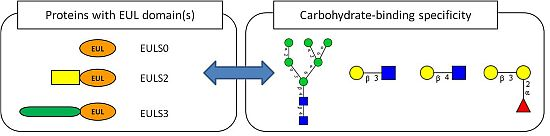
2.2. Occurrence of Plant Proteins Containing an EUL Domain
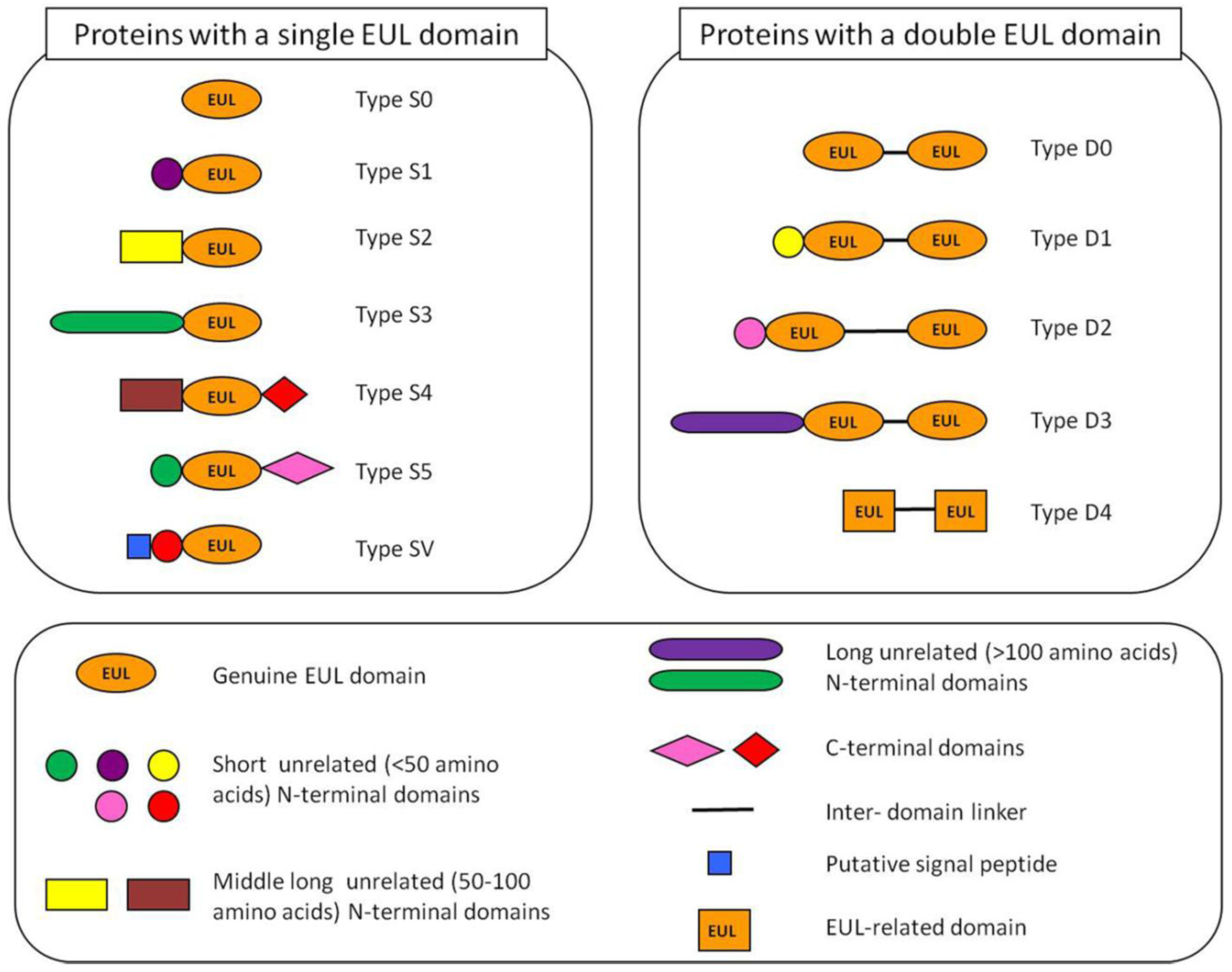
2.3. Expression of Plant Proteins Containing an EUL Domain
2.4. Analysis of EUL Proteins from Different Origins

3. Carbohydrate-Binding Properties of the EUL Domain
3.1. Agglutination and Inhibition Assays
| EEA [15,16,31] | EUL domain of OrysaEULS2 [28] | EUL domain of PhypaEULS3 | |
|---|---|---|---|
| Sugar | |||
| Mannose | +++ | +++ | - |
| Methyl α-mannopyranoside | Not tested | ++ | - |
| Galactose | - | - | ++ |
| N-acetylglucosamine | - | - | - |
| Arabinose | - | Not tested | Not tested |
| Glucose | - | - | Not tested |
| Fucose | - | Not tested | Not tested |
| Glucosamine | - | Not tested | Not tested |
| Lactose | ++ | ++ | ++ |
| Glycoprotein | |||
| Thyroglobulin | +++ | +++ | + |
| Ovomucoid | + | ++ | - |
| Asialomucin | + | +++ | ++ |
| Mucin | - | - | ++ |
| Fetuin | + | - | - |
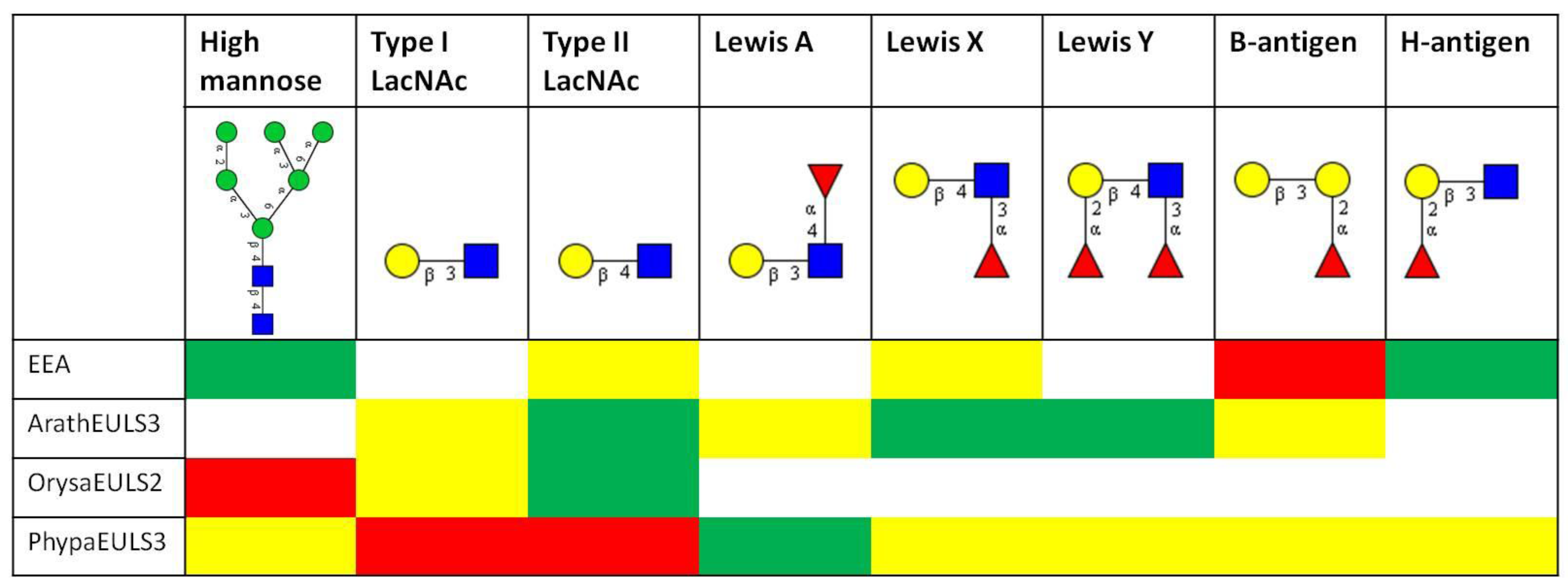
3.2. Glycan Array Analysis
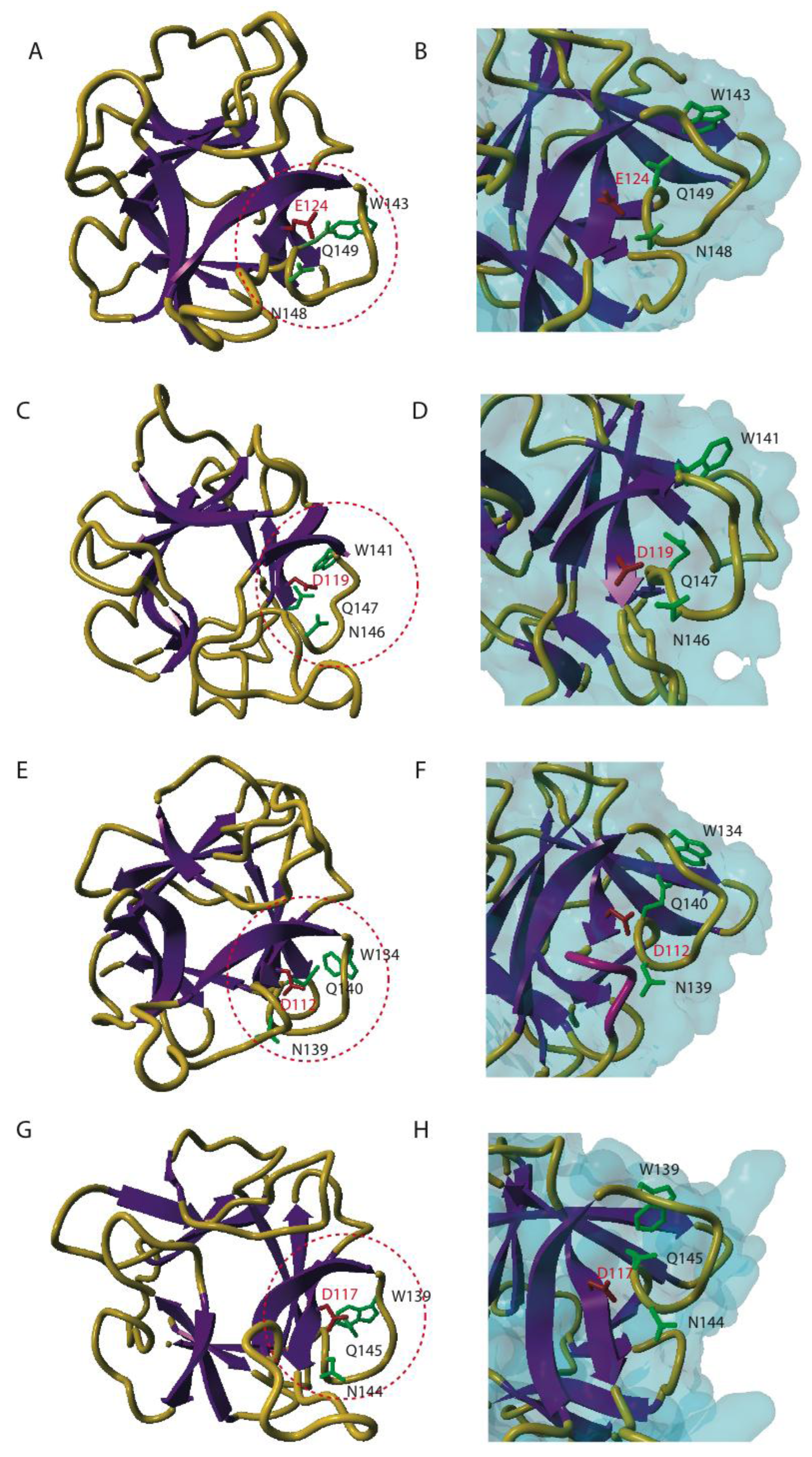
4. Three-dimensional Conformation of EUL Domains
5. Discussion
5.1. Promiscuity of the EUL Binding Site
5.2. Physiological Relevance of Carbohydrate-Binding Activity of EUL Lectins
6. Conclusions
Acknowledgments
References
- Van Damme, E.J.M.; Peumans, W.J.; Barre, A.; Rougé, P. Plant lectins: A composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit. Rev. Plant Sci. 1998, 17, 575–692. [Google Scholar]
- Peumans, W.J.; Barre, A.; Hao, Q.; Rougé, P.; Van Damme, E.J.M. Higher plants developed structurally different motifs to recognize foreign glycans. Trends Glycosci. Glycotechnol. 2000, 12, 83–101. [Google Scholar] [CrossRef]
- Vandenborre, G.; Smagghe, G.; Van Damme, E.J.M. Plant lectins as defense proteins against phytophagous insects. Phytochemistry 2011, 72, 1538–1550. [Google Scholar]
- Peumans, W.J.; Van Damme, E.J.M. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar]
- Fouquaert, E.; Peumans, W.J.; Vandekerckhove, T.T.M.; Ongenaert, M.; Van Damme, E.J.M. Proteins with an Euonymus lectin-like domain are ubiquitous in Embryophyta. BMC Plant Biol. 2009, 9, 136. [Google Scholar] [CrossRef]
- Lannoo, N.; Vandenborre, G.; Miersch, O.; Smagghe, G.; Wasternack, C.; Peumans, W.J.; Van Damme, E.J.M. The jasmonate-induced expression of the Nicotiana tabacum leaf lectin. Plant Cell Physiol. 2007, 48, 1207–1218. [Google Scholar] [CrossRef]
- Vandenborre, G.; Miersch, O.; Hause, B.; Smagghe, G.; Wasternack, C.; Van Damme, E.J.M. Spodoptera littoralis-induced lectin expression in tobacco. Plant Cell Physiol. 2009, 50, 1142–1155. [Google Scholar] [CrossRef]
- Lannoo, N.; Van Damme, E.J.M. Nucleocytoplasmic plant lectins. Biochim. Biophys. Acta 1800, 190–201. [Google Scholar]
- Van Damme, E.J.M.; Barre, A.; Rougé, P.; Peumans, W.J. Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci. 2004, 9, 484–489. [Google Scholar] [CrossRef]
- Van Damme, E.J.M.; Lannoo, N.; Fouquaert, E.; Peumans, W.J. The identification of inducible cytoplasmic/nuclear carbohydrate-binding proteins urges to develop novel concepts about the role of plant lectins. Glycoconjugate J. 2004, 20, 449–460. [Google Scholar]
- Van Damme, E.J.M.; Fouquaert, E.; Lannoo, N.; Vandenborre, G.; Schouppe, D.; Peumans, W.J. Novel concepts about the role of lectins in the plant cell. In The Molecular Immunology of Complex Carbohydrates-3; Wu, A.M., Ed.; Springer: New York, USA, 2011; Volume 705, pp. 295–324. [Google Scholar]
- Lannoo, N.; Peumans, W.J.; Van Pamel, E.; Alvarez, R.; Xiong, T.C.; Hause, G.; Mazars, C.; Van Damme, E.J.M. Localization and in vitro binding studies suggest that the cytoplasmic/nuclear tobacco lectin can interact in situ with high-mannose and complex N-glycans. FEBS Lett. 2006, 580, 6329–6337. [Google Scholar] [CrossRef]
- Schouppe, D.; Ghesquière, B.; Menschaert, G.; De Vos, W.H.; Bourque, S.; Trooskens, G.; Proost, P.; Gevaert, K.; Van Damme, E.J.M. Interaction of the tobacco lectin with histone proteins. Plant Physiol. 2011, 155, 1091–1102. [Google Scholar] [CrossRef]
- Fouquaert, E.; Peumans, W.J.; Smith, D.F.; Proost, P.; Savvides, S.N.; Van Damme, E.J.M. The “old” Euonymus europaeus agglutinin represents a novel family of ubiquitous plant proteins. Plant Physiol. 2008, 147, 1316–1324. [Google Scholar] [CrossRef]
- Pacak, F.; Kocourek, J. Studies on Phytohemagglutinins. XXV. Isolation and characterization of hemagglutinins of the spindle tree seeds (Evonymus europaea L.). Biochim. Biophys. Acta 1975, 400, 374–386. [Google Scholar] [CrossRef]
- Petryniak, J.; Pereira, M.E.; Kabat, E.A. The lectin of Euonymus europeus: purification, characterization, and an immunochemical study of its combining sit. Arch. Biochem. Biophys. 1977, 178, 118–134. [Google Scholar] [CrossRef]
- Petryniak, J.; Goldstein, I.J. Evonymus europaea lectin. Methods Enzymol. 1987, 138, 552–561. [Google Scholar]
- Van Hove, J.; Fouquaert, E.; Smith, D.F.; Proost, P.; Van Damme, E.J.M. Lectin activity of the nucleocytoplasmic EUL protein from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2011, 414, 101–105. [Google Scholar] [CrossRef]
- Unpublished data
- Moons, A.; Gielen, J.; Vandekerckhove, J.; Van der Straeten, D.; Gheysen, G.; Van Montagu, M. An abscisic-acid- and salt-stress-responsive rice cDNA from a novel plant gene family. Planta 1997, 202, 443–454. [Google Scholar] [CrossRef]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 2001, 13, 889–905. [Google Scholar]
- Dooki, A.D.; Mayer-Posner, F.J.; Askari, H.; Zaiee, A.A.; Salekdeh, G.H. Proteomic responses of rice young panicles to salinity. Proteomics 2006, 6, 6498–6507. [Google Scholar] [CrossRef]
- Ke, Y.; Han, G.; He, H.; Li, J. Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 2009, 379, 133–138. [Google Scholar] [CrossRef]
- Riccardi, F.; Gazeau, P.; de Vienne, D.; Zivy, M. Protein changes in response to progressive water deficit in maize. Plant Physiol. 1998, 117, 1253–1263. [Google Scholar] [CrossRef]
- Riccardi, F.; Gazeau, P.; Jacquemot, M.P.; Vincent, D.; Zivy, M. Deciphering genetic variations of proteome responses to water deficit in maize leaves. Plant Physiol. Biochem. 2004, 42, 1003–1011. [Google Scholar] [CrossRef]
- Samyn, B.; Sergeant, K.; Carpentier, S.; Debyser, G.; Panis, B.; Swennen, R.; Van Beeumen, J. Functional proteome analysis of the banana plant (Musa spp.) using de novo sequence analysis of derivatized peptides. J. Proteome Res. 2007, 6, 70–80. [Google Scholar] [CrossRef]
- Carpentier, S.C.; Witters, E.; Laukens, K.; Van Onckelen, H.; Swennen, R.; Panis, B. Banana (Musa spp.) as a model to study the meristem proteome: acclimation to osmotic stress. Proteomics 2007, 7, 92–105. [Google Scholar] [CrossRef]
- Al Atalah, B.; Rougé, P.; Smith, D.F.; Proost, P.; Lasanajak, Y.; Van Damme, E.J.M. Expression analysis of a type S2 EUL-related lectin from rice in Pichia pastoris. Glycoconjugate J. 2012, 29, 467–479. [Google Scholar] [CrossRef]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.E.; Alvarez, R.; Bryan, M.C.; Fazio, F.; Calarese, D.; Stevens, J.; Razi, N.; Stevens, D.J.; Skehel, J.J.; van Die, I.; Burton, D.R.; Wilson, I.A.; Cummings, R.; Bovin, N.; Wong, C.H.; Paulson, J.C. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 17033. [Google Scholar]
- Paulson, J.C.; Blixt, O.; Collins, B.E. Sweet spots in functional glycomics. Nat. Chem. Biol. 2006, 2, 238. [Google Scholar]
- Van den Bergh, K. Carbohydrate-binding proteins from spindle tree (Euonymus europaeus L.). PhD thesis, University of Leuven, Leuven, Belgium, 2003. [Google Scholar]
- Petryniak, J.; Goldstein, I.J. Immunochemical studies on the interaction between synthetic glycoconjugates and α-L-fucosyl binding lectins. Biochemistry 1986, 25, 2829–2838. [Google Scholar] [CrossRef]
- Teneberg, S.; Alsén, B.; Angström, J.; Winter, H.C.; Goldstein, I.J. Studies on Galα3-binding proteins: comparison of the glycosphingolipid binding specificities of Marasmius oreades lectin and Euonymus europaeus lectin. Glycobiology 2003, 13, 479–486. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA - a self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Arndt, J.W.; Gu, J.; Jaroszewski, L.; Schwarzenbacher, R.; Hanson, M.A.; Lebeda, F.J.; Stevens, R.C. The structure of the neurotoxin-associated protein HA33/A from Clostridium botulinum suggests a reoccurring ß-trefoil fold in the progenitor toxin complex. J. Mol. Biol. 2005, 346, 1083–1093. [Google Scholar] [CrossRef]
- Treiber, N.; Reinert, D.J.; Carpusca, I.; Aktories, K.; Schulz, G.E. Structure and mode of action of a mosquitocidal holotoxin. J. Mol. Biol. 2008, 381, 150–159. [Google Scholar] [CrossRef]
- Sulzenbacher, G.; Roig-Zamboni, V.; Peumans, W.J.; Rougé, P.; Van Damme, E.J.M.; Bourne, Y. Crystal structure of the GalNAc/Gal-specific agglutinin from the phytopathogenic ascomycete Sclerotinia sclerotiorum reveals novel adaptation of a β-trefoil domain. J. Mol. Biol. 2010, 400, 715–723. [Google Scholar] [CrossRef]
- Broom, A.; Doxey, A.C.; Lobsanov, Y.D.; Berthin, L.G.; Rose, D.R.; Howell, P.L.; McConkey, B.J.; Meiering, E.M. Modular evolution and the origins of symmetry: reconstruction of a three-fold symmetric globular protein. Structure 2012, 20, 161–171. [Google Scholar]
- Van Damme, E.J.M.; Rougé, P.; Peumans, W.J. Carbohydrate-protein interactions: Plant lectins. In Comprehensive Glycoscience-From Chemistry to Systems Biology; Kamerling, J.P., Boons, G.J., Lee, Y.C., Suzuki, A., Taniguchi, N., Voragen, A.J.G., Eds.; Elsevier: Oxford, UK, 2007; Volume 3, pp. 563–599. [Google Scholar]
- Loris, R.; De Greve, H.; Dao-Thi, M.H.; Messens, J.; Imberty, A.; Wyns, L. Structural basis of carbohydrate recognition by lectin II from Ulex europaeus, a protein with a promiscuous carbohydrate-binding site. J. Mol. Biol. 2000, 301, 987–1002. [Google Scholar] [CrossRef]
- Loris, R.; Hamelryck, T.; Bouckaert, J.; Wyns, L. Legume lectin structure. Biochim. Biophys. Acta 1998, 1383, 9–36. [Google Scholar]
- Fouquaert, E.; Smith, D.F.; Peumans, W.J.; Proost, P.; Balzarini, J.; Savvides, S.N.; Van Damme, E.J.M. Related lectins from snowdrop and maize differ in their carbohydrate-binding specificity. Biochem. Biophys. Res. Commun. 2009, 380, 260–265. [Google Scholar] [CrossRef]
- Stefanowicz, K.; Lannoo, N.; Proost, P.; Van Damme, E.J.M. Arabidopsis F-box protein containing a Nictaba-related lectin domain interacts with N-acetyllactosamine structures. FEBS Open Bio. 2012, 2, 151–158. [Google Scholar] [CrossRef]
- Nakamura, K.; Inoue, M.; Yoshiie, T.; Hosoi, K.; Kimura, Y. Changes in structural features of free N-glycan and endoglycosidase activity during tomato fruit ripening. Biosci. Biotechnol. Biochem. 2008, 72, 2936–2945. [Google Scholar] [CrossRef]
- Priem, B.; Gitti, R.; Bush, C.A.; Gross, K.C. Structure of ten free N-glycans in ripening tomato fruit. Plant Physiol. 1993, 102, 445–458. [Google Scholar]
- Fettke, J.; Hejazi, M.; Smirnova, J.; Höchel, E.; Stage, M.; Steup, M. Eukaryotic starch degradation: integration of plastidial and cytosolic pathways. J. Exp. Bot. 2009, 60, 2907–2922. [Google Scholar] [CrossRef]
- Fettke, J.; Nunes-Nesi, A.; Fernie, A.R.; Steup, M. Identification of a novel heteroglycan-interacting protein, HIP 1.3, from Arabidopsis thaliana. J. Plant Physiol. 2011, 168, 1415–1425. [Google Scholar] [CrossRef]
- Faugeron, C.; Lhernould, S.; Lemoine, J.; Costa, G.; Morvan, H. Identification of unconjugated N-glycans in strawberry plants. Plant Physiol. Biochem. 1997, 35, 891–895. [Google Scholar]
- Kimura, Y.; Takagi, S.; Shiraishi, T. Occurrence of free N-glycans in pea (Pisum sativum L) seedlings. Biosci. Biotechnol. Biochem. 1997, 61, 924–926. [Google Scholar] [CrossRef]
- Kimura, Y.; Matsuo, S. Free N-glycans already occur at an early stage of seed development. J. Biochem. 2000, 127, 1013–1019. [Google Scholar] [CrossRef]
- Kimura, Y.; Kitahara, E. Structural analysis of free N-glycans occurring in soybean seedlings. Biosci. Biotechnol. Biochem. 2000, 64, 2109–2120. [Google Scholar] [CrossRef]
- Kimura, Y.; Matsuo, S.; Tsurusaki, S.; Kimura, M.; Hara-Nishimura, I.; Nishimura, M. Subcellular localization of endo-ß-N-acetylglucosaminidase and high-mannose type free N-glycans in plant cell. Biochim. Biophys. Acta 1570, 38–46. [Google Scholar]
- Stowell, S.R.; Arthur, C.M.; Mehta, P.; Slanina, K.A.; Blixt, O.; Leffler, H.; Smith, D.F.; Cummings, R.D. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J. Biol. Chem. 2008, 283, 10109–10123. [Google Scholar]
- Liu, F.T.; Patterson, R.J.; Wang, J.L. Intracellular functions of galectins. Biochim. Biophys. Acta 2002, 1572, 263–273. [Google Scholar] [CrossRef]
- Léonard, R.; Costa, G.; Darrambide, E.; Lhernould, S.; Fleurat-Lessard, P.; Carlué, M.; Gomord, V.; Faye, L.; Maftah, A. The presence of Lewis a epitopes in Arabidopsis thaliana glycoconjugates depends on an active α4-fucosyltransferase gene. Glycobiology 2002, 12, 299–306. [Google Scholar] [CrossRef]
- Melo, N.S.; Nimtz, M.; Conradt, H.S.; Fevereiro, P.S.; Costa, J. Identification of the human Lewisa carbohydrate motif in a secretory peroxidase from a plant cell suspension culture (Vaccinium myrtillus L.). FEBS Lett. 1997, 415, 186–191. [Google Scholar] [CrossRef]
- Dam, T.K.; Cavada, B.S.; Nagano, C.S.; Rocha, B.A.M.; Benevides, R.G.; Nascimento, K.S.; de Sousa, L.A.G.; Oscarson, S.; Brewer, C.F. Fine specificities of two lectins from Cymbosema roseum seeds: a lectin specific for high-mannose oligosaccharides and a lectin specific for blood group H type II trisaccharide. Glycobiology 2011, 21, 925–933. [Google Scholar] [CrossRef]
- Kimura, Y.; Kamamoto, M.; Maeda, M.; Okano, M.; Yokoyama, M.; Kino, K. Occurrence of Lewis a epitope in N-glycans of a glycoallergen Jun a, from mountain ceder pollen. Biosci. Biotechnol. Biochem. 2005, 69, 137–144. [Google Scholar] [CrossRef]
- Maeda, M.; Kamamota, M.; Hino, K.; Yamamoto, S.; Kimura, M.; Okano, M.; Kimura, Y. Glycoformanalysis of Japanese cedar pollen allergen, Cry j 1. Biosci. Biotechnol. Biochem. 2005, 69, 1700–1705. [Google Scholar] [CrossRef]
- Maeda, M.; Kimura, Y. Glycoform analysis of N-glycans linked to glycoproteins expressed in rice culture cells: predominant occurrence of complex type N-glycans. Biosci. Biotechnol. Biochem. 2006, 70, 1356–1363. [Google Scholar] [CrossRef]
- Priem, B.; Kwan, J.S.; Wieruszeski, J.M.; Strecker, G.; Nazih, H.; Morvan, H. Isolation and characterization of free glycans of the oligomannoside type from the extracellular medium of a plant cell suspension. Glycoconjugate J. 1990, 7, 121–132. [Google Scholar] [CrossRef]
- Maeda, M.; Kimura, M.; Kimura, Y. Intracellular and extracellular free N-glycans produced by plant cells: occurrence of unusual plant complex type free N-glycans in extracellular spaces. J. Biochem. 2010, 148, 681–692. [Google Scholar] [CrossRef]
- Fitchette-Lainé, A.C.; Gomord, V.; Cabanes, M.; Michalski, J.C.; Saint Macary, M.; Foucher, B.; Cavelier, B.; Hawes, C.; Lerouge, P.; Faye, L. N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997, 12, 1411–1417. [Google Scholar]
- Moons, A.; Bauw, G.; Prinsen, E.; Van Montagu, M.; Van der Straeten, D. Molecular and physiological responses to abscisic acid and salts in roots of salt-sensitive and salt-tolerant Indica rice varieties. Plant Physiol. 1995, 107, 177–186. [Google Scholar]
- Peumans, W.J.; Winter, H.C.; Bemer, V.; Van Leuven, F.; Goldstein, I.J.; Truffa-Bachi, P.; Van Damme, E.J.M. Isolation of a novel plant lectin with an unusual specificity from Calystegia sepium. Glycoconj. J. 1997, 14, 259–265. [Google Scholar] [CrossRef]
- Peumans, W.J.; Hause, B.; Van Damme, E.J.M. The galactose-binding and mannose-binding jacalin-related lectins are located in different sub-cellular compartments. FEBS Lett. 2000, 477, 186–192. [Google Scholar] [CrossRef]
- Varki, A.; Cumming, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Marth, J.D.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Symbol nomenclature for glycan representation. Proteomics 2009, 9, 5398–5399. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fouquaert, E.; Van Damme, E.J.M. Promiscuity of the Euonymus Carbohydrate-Binding Domain. Biomolecules 2012, 2, 415-434. https://doi.org/10.3390/biom2040415
Fouquaert E, Van Damme EJM. Promiscuity of the Euonymus Carbohydrate-Binding Domain. Biomolecules. 2012; 2(4):415-434. https://doi.org/10.3390/biom2040415
Chicago/Turabian StyleFouquaert, Elke, and Els J.M. Van Damme. 2012. "Promiscuity of the Euonymus Carbohydrate-Binding Domain" Biomolecules 2, no. 4: 415-434. https://doi.org/10.3390/biom2040415