Differential Expression of 26S Proteasome Subunits and Functional Activity during Neonatal Development
Abstract
:1. Introduction
2. Results and Discussion
2.1. CTL-, TRP-, and PGPH-Like Activity Increases from Birth to Post-Weaning

2.2. Core Proteasome Subunit Expression Increases from Birth to Post-Weaning
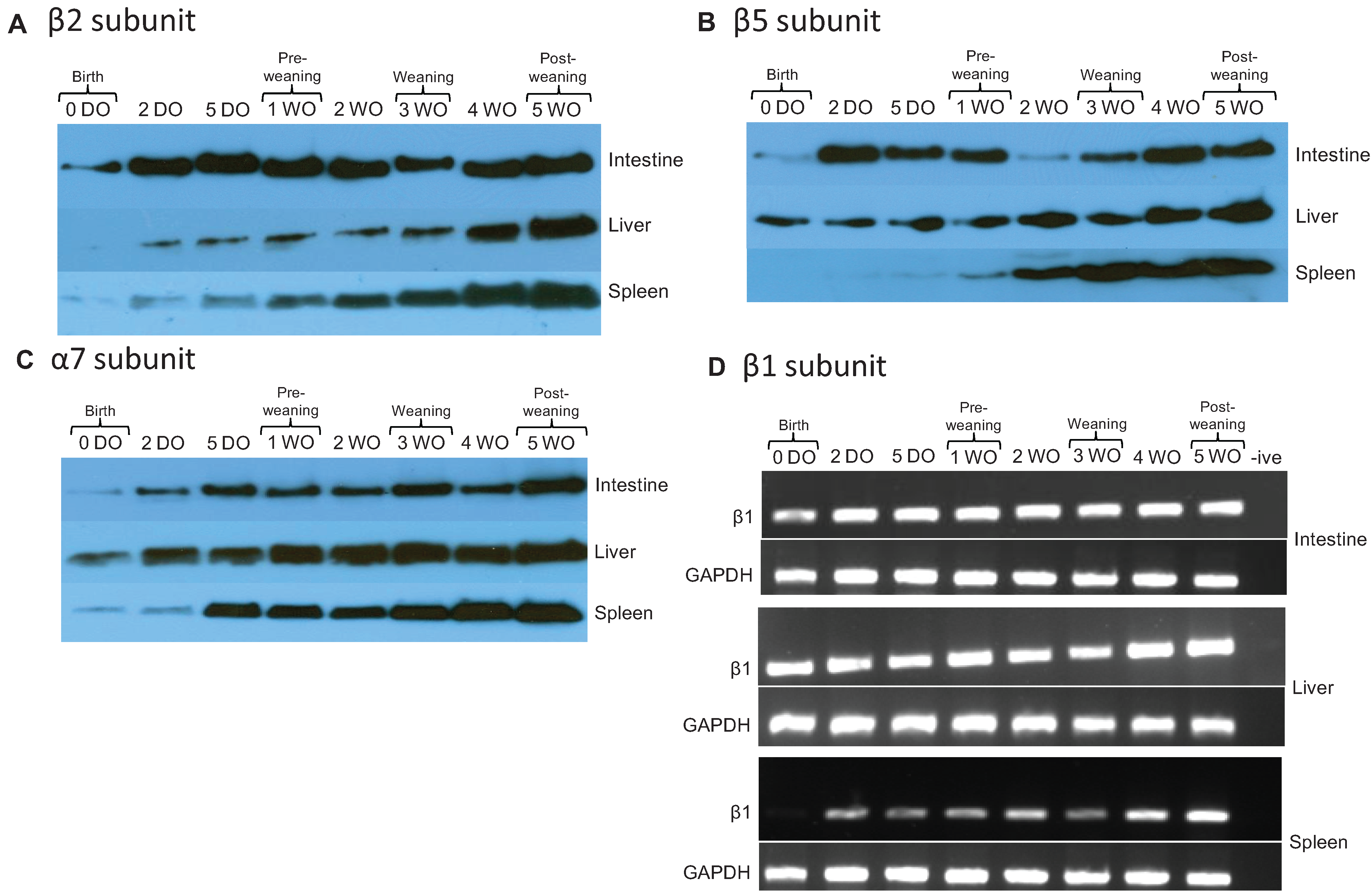
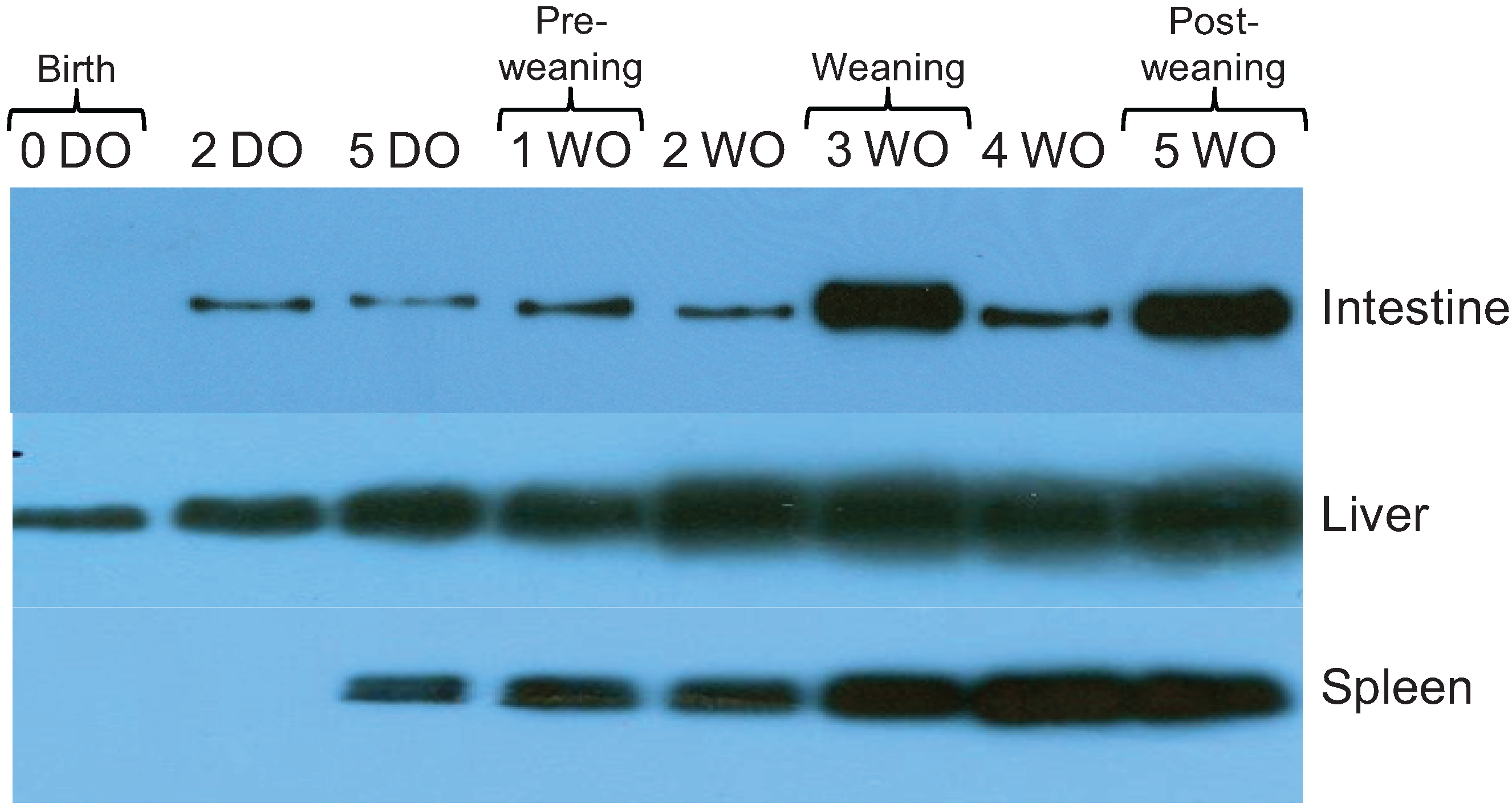
2.3. Expression of 19S Proteasome ATPase Subunit in Rat Intestine, Liver, and Spleen Increases over Time
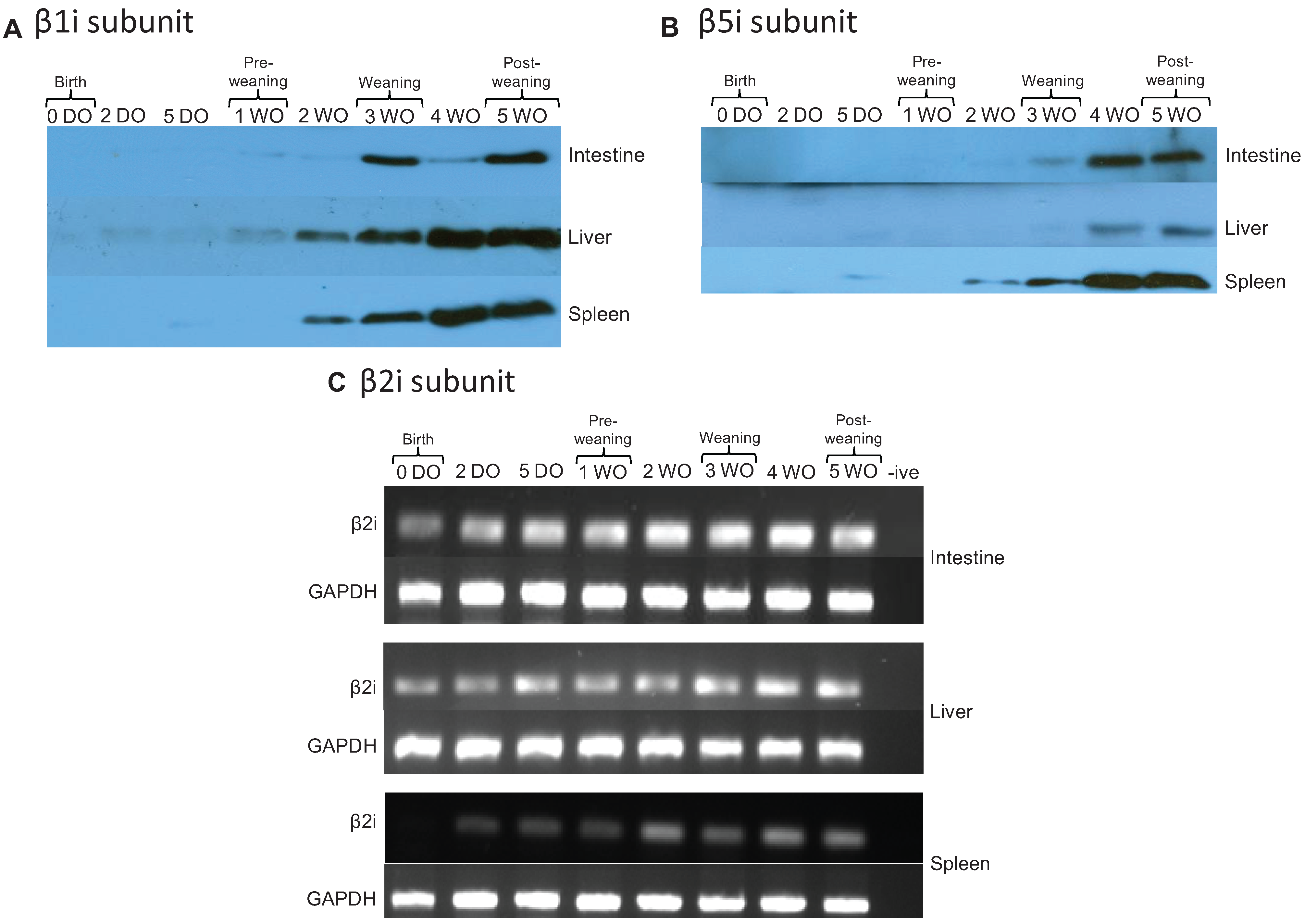
2.4. Immunoproteasomes are Developmentally Regulated and Appear at Later Time Points
3. Experimental
3.1. Ethics Statement
3.2. Animals
3.3. Antibodies, Chemicals and DNA Constructs
3.4. Expression and Purification of pET15b-mUIM-2
3.5. Expression and Purification of pGEX-2T-UBL
3.6. 26S Proteasome Purification from Rat Tissues
3.7. Proteasome Activity Assay
3.8. Western Blot Analysis
3.9. RNA Extraction and RT-PCR
3.10. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Claud, E.C.; Savidge, T.; Walker, W.A. Modulation of human intestinal epithelial cell IL-8 secretion by human milk factors. Pediatr. Res. 2003, 53, 419–425. [Google Scholar] [CrossRef]
- Nanthakumar, N.N.; Fusunyan, R.D.; Sanderson, I.; Walker, W.A. Inflammation in the developing human intestine: A possible pathophysiologic contribution to necrotizing enterocolitis. Proc. Natl. Acad. Sci. USA 2000, 97, 6043–6048. [Google Scholar] [CrossRef]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef]
- Abramova, E.B.; Sharova, N.P.; Karpov, V.L. The proteasome: destroy to live. Mol. Biol. 2002, 36, 761–776. [Google Scholar]
- Fietta, P.; Delsante, G. Proteasomes and immunoproteasomes. Riv. Biol. 2010, 103, 29–50. [Google Scholar]
- Dahlmann, B.; Ruppert, T.; Kuehn, L.; Merforth, S.; Kloetzel, P.M. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J. Mol. Biol. 2000, 303, 643–653. [Google Scholar] [CrossRef]
- Drews, O.; Wildgruber, R.; Zong, C.; Sukop, U.; Nissum, M.; Weber, G.; Gomes, A.V.; Ping, P. Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol. Cell. Proteomics 2007, 6, 2021–2031. [Google Scholar] [CrossRef]
- Kloetzel, P.M.; Ossendorp, F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 2004, 16, 76–81. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Pickart, C.M.; Cohen, R.E. Proteasomes and their kin: Proteases in the machine age. Nat. Rev. Mol. Cell Biol. 2004, 5, 177–187. [Google Scholar] [CrossRef]
- Davies, K.J. Degradation of oxidized proteins by the 20S proteasome. Biochimie 2001, 83, 301–310. [Google Scholar] [CrossRef]
- Pickering, A.M.; Davies, K.J. Degradation of damaged proteins: The main function of the 20S proteasome. Prog. Mol. Biol. Transl. Sci. 2012, 109, 227–248. [Google Scholar]
- Traenckner, E.B.; Wilk, S.; Baeuerle, P.A. A proteasome inhibitor prevents activation of NF-kappa B and stabilizes a newly phosphorylated form of I kappa B-alpha that is still bound to NF-kappa B. EMBO J. 1994, 13, 5433–5441. [Google Scholar]
- Petrof, E.O.; Kojima, K.; Ropeleski, M.J.; Musch, M.W.; Tao, Y.; de Simone, C.; Chang, E.B. Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology 2004, 127, 1474–1487. [Google Scholar] [CrossRef]
- Khan, S.; van den Broek, M.; Schwarz, K.; de Giuli, R.; Diener, P.A.; Groettrup, M. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 2001, 167, 6859–6868. [Google Scholar] [CrossRef]
- Groettrup, M.; Kirk, C.J.; Basler, M. Proteasomes in immune cells: More than peptide producers? Nat. Rev. Immunol. 2010, 10, 73–78. [Google Scholar]
- Brooks, P.; Murray, R.Z.; Mason, G.G.; Hendil, K.B.; Rivett, A.J. Association of immunoproteasomes with the endoplasmic reticulum. Biochem. J. 2000, 352, 611–615. [Google Scholar] [CrossRef]
- Gaczynska, M.; Rock, K.L.; Goldberg, A.L. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 1993, 365, 264–267. [Google Scholar] [CrossRef]
- Kremer, M.; Henn, A.; Kolb, C.; Basler, M.; Moebius, J.; Guillaume, B.; Leist, M.; van den Eynde, B.J.; Groettrup, M. Reduced immunoproteasome formation and accumulation of immunoproteasomal precursors in the brains of lymphocytic choriomeningitis virus-infected mice. J. Immunol. 2010, 185, 5549–5560. [Google Scholar] [CrossRef]
- Shibatani, T.; Nazir, M.; Ward, W.F. Alteration of rat liver 20S proteasome activities by age and food restriction. J. Gerontol. A Biol. Sci. Med. Sci. 1996, 51, B316–B322. [Google Scholar] [CrossRef]
- Noda, C.; Tanahashi, N.; Shimbara, N.; Hendil, K.B.; Tanaka, K. Tissue distribution of constitutive proteasomes, immunoproteasomes, and PA28 in rats. Biochem. Biophys. Res. Commun. 2000, 277, 348–354. [Google Scholar] [CrossRef]
- Farout, L.; Lamare, M.C.; Cardozo, C.; Harrisson, M.; Briand, Y.; Briand, M. Distribution of proteasomes and of the five proteolytic activities in rat tissues. Arch. Biochem. Biophys. 2000, 374, 207–212. [Google Scholar] [CrossRef]
- Chondrogianni, N.; Gonos, E.S. Proteasome dysfunction in mammalian aging: Steps and factors involved. Exp. Gerontol. 2005, 40, 931–938. [Google Scholar] [CrossRef]
- Visekruna, A.; Joeris, T.; Seidel, D.; Kroesen, A.; Loddenkemper, C.; Zeitz, M.; Kaufmann, S.H.; Schmidt-Ullrich, R.; Steinhoff, U. Proteasome-mediated degradation of IκBα and processing of p105 in Crohn disease and ulcerative colitis. J. Clin. Invest. 2006, 116, 3195–3203. [Google Scholar] [CrossRef]
- Elahi, S.; Ertelt, J.M.; Kinder, J.M.; Jiang, T.T.; Zhang, X.; Xin, L.; Chaturvedi, V.; Strong, B.S.; Qualls, J.E.; Steinbrecher, K.A.; et al. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature 2013, 504, 158–162. [Google Scholar] [CrossRef]
- Abramova, E.B.; Astakhova, T.M.; Sharova, N.P. Changes in proteasome activity and subunit composition during postnatal development of rat. Ontogenez 2005, 36, 205–210. [Google Scholar]
- Chen, Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell. Biol. 2005, 7, 758–765. [Google Scholar] [CrossRef]
- Hunter, C.J.; Upperman, J.S.; Ford, H.R.; Camerini, V. Understanding the susceptibility of the premature infant to necrotizing enterocolitis (NEC). Pediatr. Res. 2008, 63, 117–123. [Google Scholar] [CrossRef]
- Groettrup, M.; Soza, A.; Kuckelkorn, U.; Kloetzel, P.M. Peptide antigen production by the proteasome: Complexity provides efficiency. Immunol. Today 1996, 17, 429–435. [Google Scholar] [CrossRef]
- Sijts, E.J.; Kloetzel, P.M. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell. Mol. Life Sci. 2011, 68, 1491–1502. [Google Scholar] [CrossRef]
- Kuckelkorn, U.; Ruppert, T.; Strehl, B.; Jungblut, P.R.; Zimny-Arndt, U.; Lamer, S.; Prinz, I.; Drung, I.; Kloetzel, P.M.; Kaufmann, S.H.; et al. Link between organ-specific antigen processing by 20S proteasomes and CD8+ T cell-mediated autoimmunity. J. Exp. Med. 2002, 195, 983–990. [Google Scholar] [CrossRef]
- Chassin, C.; Kocur, M.; Pott, J.; Duerr, C.U.; Gutle, D.; Lotz, M.; Hornef, M.W. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell. Host Microbe 2010, 8, 358–368. [Google Scholar] [CrossRef]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell. 2007, 27, 731–744. [Google Scholar] [CrossRef]
- Kruger, E.; Kuckelkorn, U.; Sijts, A.; Kloetzel, P.M. The components of the proteasome system and their role in MHC class I antigen processing. Rev. Physiol. Biochem. Pharmacol. 2003, 148, 81–104. [Google Scholar]
- Wang, X.H.; Zhang, L.; Mitch, W.E.; LeDoux, J.M.; Hu, J.; Du, J. Caspase-3 cleaves specific 19S proteasome subunits in skeletal muscle stimulating proteasome activity. J. Biol. Chem. 2010, 285, 21249–21257. [Google Scholar] [CrossRef]
- Sharova, N.P.; Zakharova, L.A.; Astakhova, T.M.; Karpova, Y.D.; Melnikova, V.I.; Dmitrieva, S.B.; Lyupina, Y.V.; Erokhov, P.A. New approach to study of T cellular immunity development: Parallel investigation of lymphoid organ formation and changes in immune proteasome amount in rat early ontogenesis. Cell. Immunol. 2009, 256, 47–55. [Google Scholar] [CrossRef]
- Karpova, Y.D.; Lyupina, Y.V.; Astakhova, T.M.; Stepanova, A.A.; Erokhov, P.A.; Abramova, E.B.; Sharova, N.P. Immune proteasomes in the development of the rat immune system. Russ. J. Bioorg. Chem. 2013, 39, 356–365. [Google Scholar] [CrossRef]
- Scanlon, T.C.; Gottlieb, B.; Durcan, T.M.; Fon, E.A.; Beitel, L.K.; Trifiro, M.A. Isolation of human proteasomes and putative proteasome-interacting proteins using a novel affinity chromatography method. Exp. Cell. Res. 2009, 315, 176–189. [Google Scholar] [CrossRef]
- Claud, E.C.; McDonald, J.A.K.; He, S.-M.; Yu, Y.; Duong, L.; Sun, J.; Petrof, E.O. Department of Medicine, Queen’s University, Kingston, ON K7L 2V7: Canada, Unpublished data; 2014.
- Mueller, T.D.; Feigon, J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003, 22, 4634–4645. [Google Scholar] [CrossRef]
- Besche, H.C.; Haas, W.; Gygi, S.P.; Goldberg, A.L. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 2009, 48, 2538–2549. [Google Scholar] [CrossRef]
- Sijts, A.; Sun, Y.; Janek, K.; Kral, S.; Paschen, A.; Schadendorf, D.; Kloetzel, P.M. The role of the proteasome activator PA28 in MHC class I antigen processing. Mol. Immunol. 2002, 39, 165–169. [Google Scholar] [CrossRef]
- Amarri, S.; Benatti, F.; Callegari, M.L.; Shahkhalili, Y.; Chauffard, F.; Rochat, F.; Acheson, K.J.; Hager, C.; Benyacoub, J.; Galli, E.; et al. Changes of gut microbiota and immune markers during the complementary feeding period in healthy breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 488–495. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Claud, E.C.; McDonald, J.A.K.; He, S.-M.; Yu, Y.; Duong, L.; Sun, J.; Petrof, E.O. Differential Expression of 26S Proteasome Subunits and Functional Activity during Neonatal Development. Biomolecules 2014, 4, 812-826. https://doi.org/10.3390/biom4030812
Claud EC, McDonald JAK, He S-M, Yu Y, Duong L, Sun J, Petrof EO. Differential Expression of 26S Proteasome Subunits and Functional Activity during Neonatal Development. Biomolecules. 2014; 4(3):812-826. https://doi.org/10.3390/biom4030812
Chicago/Turabian StyleClaud, Erika C., Julie A. K. McDonald, Shu-Mei He, Yueyue Yu, Lily Duong, Jun Sun, and Elaine O. Petrof. 2014. "Differential Expression of 26S Proteasome Subunits and Functional Activity during Neonatal Development" Biomolecules 4, no. 3: 812-826. https://doi.org/10.3390/biom4030812



