The Immunoglobulins of Cold-Blooded Vertebrates
Abstract
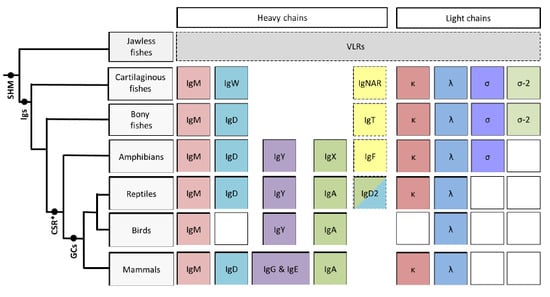
:1. Introduction
2. Cartilaginous Fishes


3. Bony Fishes

4. Amphibians
5. Reptiles
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Flajnik, M.F.; Kasahara, M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat. Rev. Genet. 2010, 11, 47–59. [Google Scholar] [CrossRef]
- Pancer, Z.; Amemiya, C.T.; Ehrhardt, G.R.; Ceitlin, J.; Gartland, G.L.; Cooper, M.D. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004, 430, 174–180. [Google Scholar] [CrossRef]
- Pancer, Z.; Saha, N.R.; Kasamatsu, J.; Suzuki, T.; Amemiya, C.T.; Kasahara, M.; Cooper, M.D. Variable lymphocyte receptors in hagfish. Proc. Natl. Acad. Sci. USA 2005, 102, 9224–9229. [Google Scholar] [CrossRef]
- Inoue, J.G.; Miya, M.; Lam, K.; Tay, B.H.; Danks, J.A.; Bell, J.; Walker, T.I.; Venkatesh, B. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): A mitogenomic perspective. Mol. Biol. Evol. 2010, 27, 2576–2586. [Google Scholar] [CrossRef]
- Dooley, H.; Flajnik, M.F. Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol. 2006, 30, 43–56. [Google Scholar] [CrossRef]
- Miracle, A.L.; Anderson, M.K.; Litman, R.T.; Walsh, C.J.; Luer, C.A.; Rothenberg, E.V.; Litman, G.W. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int. Immunol. 2001, 13, 567–580. [Google Scholar] [CrossRef]
- Zapata, A.; Amemiya, C.T. Phylogeny of lower vertebrates and their immunological structures. Curr. Top. Microbiol. Immunol. 2000, 248, 67–107. [Google Scholar]
- Hinds, K.R.; Litman, G.W. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature 1986, 320, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Malecek, K.; Lee, V.; Feng, W.; Huang, J.L.; Flajnik, M.F.; Ohta, Y.; Hsu, E. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol. 2008, 6, e157. [Google Scholar] [PubMed]
- Kokubu, F.; Litman, R.; Shamblott, M.J.; Hinds, K.; Litman, G.W. Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J. 1988, 7, 3413–3422. [Google Scholar] [PubMed]
- Lee, S.S.; Fitch, D.; Flajnik, M.F.; Hsu, E. Rearrangement of immunoglobulin genes in shark germ cells. J. Exp. Med. 2000, 191, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Stanfield, R.L.; Greenberg, A.S.; Flajnik, M.F. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): Identification of a new locus preferentially expressed in early development. Immunogenetics 2002, 54, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Rumfelt, L.L.; Avila, D.; Diaz, M.; Bartl, S.; McKinney, E.C.; Flajnik, M.F. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc. Natl. Acad. Sci. USA 2001, 98, 1775–1780. [Google Scholar] [CrossRef] [PubMed]
- Rogozin, I.B.; Iyer, L.M.; Liang, L.; Glazko, G.V.; Liston, V.G.; Pavlov, Y.I.; Aravind, L.; Pancer, Z. Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 2007, 8, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Warr, G.W.; Magor, K.E.; Higgins, D.A. IgY: Clues to the origins of modern antibodies. Immunol. Today 1995, 16, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Mashoof, S.; Goodroe, A.; Du, C.C.; Eubanks, J.O.; Jacobs, N.; Steiner, J.M.; Tizard, I.; Suchodolski, J.S.; Criscitiello, M.F. Ancient T-independence of mucosal IgX/A: Gut microbiota unaffected by larval thymectomy in Xenopus laevis. Mucosal Immunol. 2013, 6, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Saha, N.R.; Ota, T.; Litman, G.W.; Hansen, J.; Parra, Z.; Hsu, E.; Buonocore, F.; Canapa, A.; Cheng, J.F.; Amemiya, C.T.; et al. Genome complexity in the coelacanth is reflected in its adaptive immune system. J. Exp. Zool. B Mol. Dev. Evol. 2014, 322, 438–463. [Google Scholar] [CrossRef] [PubMed]
- Criscitiello, M.F.; Flajnik, M.F. Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur. J. Immunol. 2007, 37, 2683–2694. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Lee, V.; Finn, A.; Senger, K.; Zarrin, A.A.; du Pasquier, L.; Hsu, E. Origin of immunoglobulin isotype switching. Curr. Biol. 2012, 22, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Saunders, H.L.; Oko, A.L.; Scott, A.N.; Fan, C.W.; Magor, B.G. The cellular context of AID expressing cells in fish lymphoid tissues. Dev. Comp. Immunol. 2010, 34, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Marchalonis, J.; Edelman, G.M. Polypeptide chains of immunoglobulins from the smooth dogfish (Mustelus canis). Science 1966, 154, 1567–1568. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Flajnik, M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc. Natl. Acad. Sci. USA 2006, 103, 10723–10728. [Google Scholar] [CrossRef] [PubMed]
- Clem, I.W.; De, B.F.; Sigel, M.M. Phylogeny of immunoglobulin structure and function. II. Immunoglobulins of the nurse shark. J. Immunol. 1967, 99, 1226–1235. [Google Scholar] [PubMed]
- Dooley, H.; Flajnik, M.F. Shark immunity bites back: Affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur. J. Immunol. 2005, 35, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Small, P.A., Jr.; Klapper, D.G.; Clem, L.W. Half-lives, body distribution and lack of interconversion of serum 19S and 7S IgM of sharks. J. Immunol. 1970, 105, 29–37. [Google Scholar] [PubMed]
- Castro, C.D.; Ohta, Y.; Dooley, H.; Flajnik, M.F. Noncoordinate expression of J-chain and Blimp-1 define nurse shark plasma cell populations during ontogeny. Eur. J. Immunol. 2013, 43, 3061–3075. [Google Scholar] [CrossRef] [PubMed]
- Hohman, V.S.; Stewart, S.E.; Rumfelt, L.L.; Greenberg, A.S.; Avila, D.W.; Flajnik, M.F.; Steiner, L.A. J chain in the nurse shark: Implications for function in a lower vertebrate. J. Immunol. 2003, 170, 6016–6023. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F; Hsu, E.; Department of Microbiology & Immunology, University of Maryland School of Medicine, Baltimore, MD, USA. Unpublished work. 2001.
- Greenberg, A.S.; Hughes, A.L.; Guo, J.; Avila, D.; McKinney, E.C.; Flajnik, M.F. A novel “chimeric” antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur. J. Immunol. 1996, 26, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.K.; Strong, S.J.; Litman, R.T.; Luer, C.A.; Amemiya, C.T.; Rast, J.P.; Litman, G.W. A long form of the skate IgX gene exhibits a striking resemblance to the new shark IgW and IgNARC genes. Immunogenetics 1999, 49, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Tomonaga, S. The second immunoglobulin class is commonly present in cartilaginous fish belonging to the order Rajiformes. Mol. Immunol. 1988, 25, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Rumfelt, L.L.; Diaz, M.; Lohr, R.L.; Mochon, E.; Flajnik, M.F. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol. 2004, 173, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.E.; Crouch, K.; Cao, W.; Muller, M.R.; Wu, L.; Steven, J.; Lee, M.; Liang, M.; Flajnik, M.F.; Shih, H.H.; et al. Characterization of the immunoglobulin repertoire of the spiny dogfish (Squalus acanthias). Dev. Comp. Immunol. 2012, 36, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; du Pasquier, L.; Hsu, E. Shark IgW C region diversification through RNA processing and isotype switching. J. Immunol. 2013, 191, 3410–3418. [Google Scholar] [CrossRef] [PubMed]
- Eason, D.D.; Litman, R.T.; Luer, C.A.; Kerr, W.; Litman, G.W. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur. J. Immunol. 2004, 34, 2551–2558. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, R.L.; Dooley, H.; Flajnik, M.F.; Wilson, I.A. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004, 305, 1770–1773. [Google Scholar] [CrossRef] [PubMed]
- Criscitiello, M.F.; Saltis, M.; Flajnik, M.F. An evolutionarily mobile antigen receptor variable region gene: Doubly rearranging NAR-TcR genes in sharks. Proc. Natl. Acad. Sci. USA 2006, 103, 5036–5041. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Greenberg, A.S.; Flajnik, M.F. Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: Possible role in antigen-driven reactions in the absence of germinal centers. Proc. Natl. Acad. Sci. USA 1998, 95, 14343–14348. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Fidler, J.E.; Clem, L.W.; Small, P.A., Jr. Immunoglobulin synthesis in neonatal nurse sharks (Ginglymostoma cirratum). Comp. Biochem. Physiol. 1969, 31, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Rumfelt, L.L.; McKinney, E.C.; Taylor, E.; Flajnik, M.F. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand. J. Immunol. 2002, 56, 130–148. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.; Flajnik, M.F.; School of Biological Sciences, University of Aberdeen, Aberdeen, UK. Unpublished work. 2005.
- Roux, K.H.; Greenberg, A.S.; Greene, L.; Strelets, L.; Avila, D.; McKinney, E.C.; Flajnik, M.F. Structural analysis of the nurse shark (new) antigen receptor (NAR): Molecular convergence of NAR and unusual mammalian immunoglobulins. Proc. Natl. Acad. Sci. USA 1998, 95, 11804–11809. [Google Scholar] [CrossRef] [PubMed]
- Crouch, K.; Smith, L.E.; Williams, R.; Cao, W.; Lee, M.; Jensen, A.; Dooley, H. Humoral immune response of the small-spotted catshark, Scyliorhinus canicula. Fish Shellfish Immunol. 2013, 34, 1158–1169. [Google Scholar] [PubMed]
- Dooley, H.; Flajnik, M.F.; School of Biological Sciences, University of Aberdeen, Aberdeen, UK. Unpublished work. 2004.
- Lee, S.S.; Tranchina, D.; Ohta, Y.; Flajnik, M.F.; Hsu, E. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 2002, 16, 571–582. [Google Scholar] [CrossRef]
- Diaz, M.; Velez, J.; Singh, M.; Cerny, J.; Flajnik, M.F. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: The translesion synthesis model of somatic hypermutation. Int. Immunol. 1999, 11, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F. Comparative analyses of immunoglobulin genes: Surprises and portents. Nat. Rev. Immunol. 2002, 2, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Sunyer, J.O. Fishing for mammalian paradigms in the teleost immune system. Nat. Immunol. 2013, 14, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Kaattari, S.; Evans, D.; Klemer, J. Varied redox forms of teleost IgM: An alternative to isotypic diversity? Immunol. Rev. 1998, 166, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Bromage, E.; Kaattari, I.; Kaattari, S. Transduction of binding affinity by B lymphocytes: A new dimension in immunological regulation. Dev. Comp. Immunol. 2011, 35, 982–990. [Google Scholar] [CrossRef] [PubMed]
- Cain, K.D.; Jones, D.R.; Raison, R.L. Antibody-antigen kinetics following immunization of rainbow trout (Oncorhynchus mykiss) with a T-cell dependent antigen. Dev. Comp. Immunol. 2002, 26, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Kaattari, S.L.; Zhang, H.L.; Khor, I.W.; Kaattari, I.M.; Shapiro, D.A. Affinity maturation in trout: Clonal dominance of high affinity antibodies late in the immune response. Dev. Comp. Immunol. 2002, 26, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Kaattari, I.M.; Ma, C.; Kaattari, S. The teleost humoral immune response. Fish Shellfish Immunol. 2013, 35, 1719–1728. [Google Scholar] [CrossRef]
- Wilson, M.; Bengten, E.; Miller, N.W.; Clem, L.W.; du Pasquier, L.; Warr, G.W. A novel chimeric Ig heavy chain from a teleost fish shares similarities to IgD. Proc. Natl. Acad. Sci. USA 1997, 94, 4593–4597. [Google Scholar] [CrossRef] [PubMed]
- Edholm, E.S.; Bengten, E.; Stafford, J.L.; Sahoo, M.; Taylor, E.B.; Miller, N.W.; Wilson, M. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J. Immunol. 2010, 185, 4082–4094. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Bromage, E.; Abos, B.; Pignatelli, J.; Gonzalez, G.A.; Luque, A.; Tafalla, C. CCR7 is mainly expressed in teleost gills, where it defines an IgD+IgM- B lymphocyte subset. J. Immunol. 2014, 192, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Billian, G.; Bella, C.; Mondiere, P.; Defrance, T. Identification of a tonsil IgD+ B cell subset with phenotypical and functional characteristics of germinal center B cells. Eur. J. Immunol. 1996, 26, 1712–1719. [Google Scholar] [CrossRef] [PubMed]
- Edholm, E.S.; Bengten, E.; Wilson, M. Insights into the function of IgD. Dev. Comp. Immunol. 2011, 35, 1309–1316. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gomez, F.; Greene, W.; Rego, K.; Hansen, J.D.; Costa, G.; Kataria, P.; Bromage, E.S. Discovery and characterization of secretory IgD in rainbow trout: Secretory IgD is produced through a novel splicing mechanism. J. Immunol. 2012, 188, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Xu, W.; Wilson, M.; He, B.; Miller, N.W.; Bengten, E.; Edholm, E.S.; Santini, P.A.; Rath, P.; Chiu, A.; et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat. Immunol. 2009, 10, 889–898. [Google Scholar] [PubMed]
- Hansen, J.D.; Landis, E.D.; Phillips, R.B. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc. Natl. Acad. Sci. USA 2005, 102, 6919–6924. [Google Scholar] [CrossRef] [PubMed]
- Danilova, N.; Bussmann, J.; Jekosch, K.; Steiner, L.A. The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z. Nat. Immunol. 2005, 6, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F. All GOD’s creatures got dedicated mucosal immunity. Nat. Immunol. 2010, 11, 777–779. [Google Scholar] [CrossRef] [PubMed]
- Fillatreau, S.; Six, A.; Magadan, S.; Castro, R.; Sunyer, J.O.; Boudinot, P. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front. Immunol. 2013. [Google Scholar] [CrossRef]
- Zhang, Y.A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Parra, D.; Gomez, D.; Salinas, I.; Zhang, Y.A.; von Gersdorff, J.L.; Heinecke, R.D.; Buchmann, K.; Lapatra, S.; Sunyer, J.O.; et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. USA 2013, 110, 13097–13102. [Google Scholar] [CrossRef] [PubMed]
- Ota, T.; Rast, J.P.; Litman, G.W.; Amemiya, C.T. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc. Natl. Acad. Sci. USA 2003, 100, 2501–2506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tacchi, L.; Wei, Z.; Zhao, Y.; Salinas, I. Intraclass diversification of immunoglobulin heavy chain genes in the African lungfish. Immunogenetics 2014, 66, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Edholm, E.S.; Wilson, M.; Bengten, E. Immunoglobulin light (IgL) chains in ectothermic vertebrates. Dev. Comp. Immunol. 2011, 35, 906–915. [Google Scholar] [CrossRef] [PubMed]
- Edholm, E.S.; Wilson, M.; Sahoo, M.; Miller, N.W.; Pilstrom, L.; Wermenstam, N.E.; Bengten, E. Identification of Igsigma and Iglambda in channel catfish, Ictalurus punctatus, and Iglambda in Atlantic cod, Gadus morhua. Immunogenetics 2009, 61, 353–370. [Google Scholar] [CrossRef] [PubMed]
- Solem, S.T.; Stenvik, J. Antibody repertoire development in teleosts—A review with emphasis on salmonids and Gadus morhua L. Dev. Comp. Immunol. 2006, 30, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Magnadottir, B.; Gudmundsdottir, S.; Gudmundsdottir, B.K.; Helgason, S. Natural antibodies of cod (Gadus morhua L.): Specificity, activity and affinity. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 154, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrom, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malmstrom, M.; Jentoft, S.; Gregers, T.F.; Jakobsen, K.S. Unraveling the evolution of the Atlantic cod’s (Gadus morhua L.) alternative immune strategy. PLoS One 2013, 8, e74004. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Pan-Hammarstrom, Q.; Yu, S.; Wertz, N.; Zhang, X.; Li, N.; Butler, J.E.; Hammarstrom, L. Identification of IgF, a hinge-region-containing Ig class, and IgD in Xenopus tropicalis. Proc. Natl. Acad. Sci. USA 2006, 103, 12087–12092. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Wu, Q.; Ren, L.; Hu, X.; Guo, Y.; Warr, G.W.; Hammarstrom, L.; Li, N.; Zhao, Y. Expression of IgM, IgD, and IgY in a reptile, Anolis carolinensis. J. Immunol. 2009, 183, 3858–3864. [Google Scholar] [CrossRef] [PubMed]
- Magadan-Mompo, S.; Sanchez-Espinel, C.; Gambon-Deza, F. Immunoglobulin genes of the turtles. Immunogenetics 2013, 65, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Magadan-Mompo, S.; Sanchez-Espinel, C.; Gambon-Deza, F. IgH loci of American alligator and saltwater crocodile shed light on IgA evolution. Immunogenetics 2013, 65, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Rabbani, H.; Shimizu, A.; Hammarstrom, L. Mapping of the chicken immunoglobulin heavy-chain constant region gene locus reveals an inverted alpha gene upstream of a condensed upsilon gene. Immunology 2000, 101, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.A.; Jiang, N.; White, R.A., III; Fisher, D.S.; Quake, S.R. High-throughput sequencing of the zebrafish antibody repertoire. Science 2009, 324, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Yasuike, M.; de Boer, J.; von Schalburg, K.R.; Cooper, G.A.; McKinnel, L.; Messmer, A.; So, S.; Davidson, W.S.; Koop, B.F. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC Genomics 2010. [Google Scholar] [CrossRef]
- Castro, R.; Jouneau, L.; Pham, H.P.; Bouchez, O.; Giudicelli, V.; Lefranc, M.P.; Quillet, E.; Benmansour, A.; Cazals, F.; Six, A.; et al. Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog. 2013, 9, e1003098. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Kaattari, I.M.; Kaattari, S.L. The differential dynamics of antibody subpopulation expression during affinity maturation in a teleost. Fish Shellfish Immunol. 2011, 30, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Waldbieser, G.C.; Lobb, C.J. The nucleotide targets of somatic mutation and the role of selection in immunoglobulin heavy chains of a teleost fish. J. Immunol. 2006, 176, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Pan-Hammarstrom, Q.; Zhao, Z.; Hammarstrom, L. Identification of the activation-induced cytidine deaminase gene from zebrafish: An evolutionary analysis. Dev. Comp. Immunol. 2005, 29, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Barreto, V.M.; Pan-Hammarstrom, Q.; Zhao, Y.; Hammarstrom, L.; Misulovin, Z.; Nussenzweig, M.C. AID from bony fish catalyzes class switch recombination. J. Exp. Med. 2005, 202, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Wakae, K.; Magor, B.G.; Saunders, H.; Nagaoka, H.; Kawamura, A.; Kinoshita, K.; Honjo, T.; Muramatsu, M. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int. Immunol. 2006, 18, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Steiner, L.A.; Mikoryak, C.A.; Lopes, A.D.; Green, C. Immunoglobulins in ranid frogs and tadpoles. Adv. Exp. Med. Biol. 1975, 64, 173–183. [Google Scholar] [PubMed]
- Hadji-Azimi, I. Structural studies of the Xenopus 19S immunoglobulin and 7S immunoglobulin and two immunoglobulin-like proteins. Immunology 1975, 28, 419–429. [Google Scholar] [PubMed]
- Qin, T.; Ren, L.; Hu, X.; Guo, Y.; Fei, J.; Zhu, Q.; Butler, J.E.; Wu, C.; Li, N.; Hammarstrom, L.; et al. Genomic organization of the immunoglobulin light chain gene loci in Xenopus tropicalis: Evolutionary implications. Dev. Comp. Immunol. 2008, 32, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Du Pasquier, L.; Schwager, J.; Flajnik, M.F. The immune system of Xenopus. Annu. Rev. Immunol. 1989, 7, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Hsu, E.; Flajnik, M.F.; du Pasquier, L. A third immunoglobulin class in amphibians. J. Immunol. 1985, 135, 1998–2004. [Google Scholar] [PubMed]
- Zhu, R.; Chen, Z.Y.; Wang, J.; Yuan, J.D.; Liao, X.Y.; Gui, J.F.; Zhang, Q.Y. Thymus cDNA library survey uncovers novel features of immune molecules in Chinese giant salamander Andrias davidianus. Dev. Comp. Immunol. 2014, 46, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Mussmann, R.; du Pasquier, L.; Hsu, E. Is Xenopus IgX an analog of IgA? Eur. J. Immunol. 1996, 26, 2823–2830. [Google Scholar] [CrossRef] [PubMed]
- Du, C.C.; Mashoof, S.M.; Criscitiello, M.F. Oral immunization of the African clawed frog (Xenopus laevis) upregulates the mucosal immunoglobulin IgX. Vet. Immunol. Immunopathol. 2012, 145, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Gatto, D.; Sainsbury, E.; Harriman, G.R.; Hengartner, H.; Zinkernagel, R.M. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000, 288, 2222–2226. [Google Scholar] [CrossRef] [PubMed]
- Mansikka, A. Chicken IgA H chains. Implications concerning the evolution of H chain genes. J. Immunol. 1992, 149, 855–861. [Google Scholar] [PubMed]
- Marchalonis, J.; Edelman, G.M. Phylogenetic origins of antibody structure II. Immunoglobulins in the primary immune response of the bullfrog, Rana catesbiana. J. Exp. Med. 1966, 124, 901–913. [Google Scholar] [CrossRef]
- Schaerlinger, B.; Frippiat, J.P. IgX antibodies in the urodele amphibian Ambystoma mexicanum. Dev. Comp. Immunol. 2008, 32, 908–915. [Google Scholar]
- Fellah, J.S.; Iscaki, S.; Vaerman, J.P.; Charlemagne, J. Transient developmental expression of IgY and secretory component like protein in the gut of the axolotl (Ambystoma mexicanum). Dev. Immunol. 1992, 2, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Schaerlinger, B.; Bascove, M.; Frippiat, J.P. A new isotype of immunoglobulin heavy chain in the urodele amphibian Pleurodeles waltl predominantly expressed in larvae. Mol. Immunol. 2008, 45, 776–786. [Google Scholar] [PubMed]
- Das, S.; Nikolaidis, N.; Klein, J.; Nei, M. Evolutionary redefinition of immunoglobulin light chain isotypes in tetrapods using molecular markers. Proc. Natl. Acad. Sci. USA 2008, 105, 16647–16652. [Google Scholar] [CrossRef] [PubMed]
- Mussmann, R.; Courtet, M.; Schwager, J.; du Pasquier, L. Microsites for immunoglobulin switch recombination breakpoints from Xenopus to mammals. Eur. J. Immunol. 1997, 27, 2610–2619. [Google Scholar] [CrossRef] [PubMed]
- Zarrin, A.A.; Alt, F.W.; Chaudhuri, J.; Stokes, N.; Kaushal, D.; du Pasquier, L.; Tian, M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 2004, 5, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Du Pasquier, L.; Robert, J.; Courtet, M.; Mussmann, R. B-cell development in the amphibian Xenopus. Immunol. Rev. 2000, 175, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Poorten, T.J.; Kuhn, R.E. Maternal transfer of antibodies to eggs in Xenopus laevis. Dev. Comp. Immunol. 2009, 33, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Marr, S.; Morales, H.; Bottaro, A.; Cooper, M.; Flajnik, M.; Robert, J. Localization and differential expression of activation-induced cytidine deaminase in the amphibian Xenopus upon antigen stimulation and during early development. J. Immunol. 2007, 179, 6783–6789. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.; Hsu, E.; Marcuz, A.; Courtet, M.; du Pasquier, L.; Steinberg, C. What limits affinity maturation of antibodies in Xenopus—The rate of somatic mutation or the ability to select mutants? EMBO J. 1992, 11, 4337–4347. [Google Scholar] [PubMed]
- Ramsey, J.P.; Reinert, L.K.; Harper, L.K.; Woodhams, D.C.; Rollins-Smith, L.A. Immune defenses against Batrachochytrium dendrobatidis, a fungus linked to global amphibian declines, in the South African clawed frog, Xenopus laevis. Infect. Immun. 2010, 78, 3981–3992. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.M.; Hsu, E. Abbreviated junctional sequences impoverish antibody diversity in urodele amphibians. J. Immunol. 1997, 159, 3391–3399. [Google Scholar] [PubMed]
- Frippiat, J.P. Contribution of the urodele amphibian Pleurodeles waltl to the analysis of spaceflight-associated immune system deregulation. Mol. Immunol. 2013, 56, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Janes, D.E.; Organ, C.L.; Fujita, M.K.; Shedlock, A.M.; Edwards, S.V. Genome evolution in Reptilia, the sister group of mammals. Annu. Rev. Genomics Hum. Genet. 2010, 11, 239–264. [Google Scholar] [CrossRef] [PubMed]
- Marchalonis, J.J.; Ealey, E.H.; Diener, E. Immune response of the tuatara, Sphenodon punctatum. Aust. J. Exp. Biol. Med. Sci. 1969, 47, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Gambon-Deza, F.; Sanchez-Espinel, C.; Mirete-Bachiller, S.; Magadan-Mompo, S. Snakes antibodies. Dev. Comp. Immunol. 2012, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gambon, D.F.; Sanchez, E.C.; Magadan, M.S. The immunoglobulin heavy chain locus in the reptile Anolis carolinensis. Mol. Immunol. 2009, 46, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Hirano, M.; Tako, R.; McCallister, C.; Nikolaidis, N. Evolutionary genomics of immunoglobulin-encoding Loci in vertebrates. Curr. Genomics 2012, 13, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Turchin, A.; Hsu, E. The generation of antibody diversity in the turtle. J. Immunol. 1996, 156, 3797–3805. [Google Scholar] [PubMed]
- Natarajan, K.; Muthukkaruppan, V.R. Immunoglobulin classes in the garden lizard, Calotes versicolor. Dev. Comp. Immunol. 1984, 8, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Gao, Y.; Wang, T.; Sun, Y.; Wei, Z.; Li, L.; Ren, L.; Guo, Y.; Hu, X.; Lu, Y.; et al. Extensive diversification of IgH subclass-encoding genes and IgM subclass switching in crocodilians. Nat. Commun. 2013. [Google Scholar] [CrossRef]
- Leslie, G.A.; Clem, L.W. Phylogeny of immunoglobulin structure and function. VI. 17S, 7.5S and 5.7S anti-DNP of the turtle, Pseudamys scripta. J. Immunol. 1972, 108, 1656–1664. [Google Scholar] [PubMed]
- Gambon-Deza, F.; Espinel, C.S. IgD in the reptile leopard gecko. Mol. Immunol. 2008, 45, 3470–3476. [Google Scholar] [CrossRef] [PubMed]
- Iwata, A.; Iwase, T.; Ogura, Y.; Takahashi, T.; Matsumoto, N.; Yoshida, T.; Kamei, N.; Kobayashi, K.; Mestecky, J.; Moro, I.; et al. Cloning and expression of the turtle (Trachemys scripta) immunoglobulin joining (J)-chain cDNA. Immunogenetics 2002, 54, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, T.; Sun, Y.; Cheng, G.; Yang, H.; Wei, Z.; Wang, P.; Hu, X.; Ren, L.; Meng, Q.; et al. Extensive diversification of IgD-, IgY-, and truncated IgY(deltaFc)-encoding genes in the red-eared turtle (Trachemys scripta elegans). J. Immunol. 2012, 189, 3995–4004. [Google Scholar] [CrossRef] [PubMed]
- Magor, K.E.; Higgins, D.A.; Middleton, D.L.; Warr, G.W. One gene encodes the heavy chains for three different forms of IgY in the duck. J. Immunol. 1994, 153, 5549–5555. [Google Scholar] [PubMed]
- Humphrey, B.D.; Calvert, C.C.; Klasing, K.C. The ratio of full length IgY to truncated IgY in immune complexes affects macrophage phagocytosis and the acute phase response of mallard ducks (Anas platyrhynchos). Dev. Comp. Immunol. 2004, 28, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, G.L.; Nie, P. IgM, IgD and IgY and their expression pattern in the Chinese soft-shelled turtle Pelodiscus sinensis. Mol. Immunol. 2009, 46, 2124–2132. [Google Scholar] [CrossRef] [PubMed]
- Deza, F.G.; Espinel, C.S.; Beneitez, J.V. A novel IgA-like immunoglobulin in the reptile Eublepharis macularius. Dev. Comp. Immunol. 2007, 31, 596–605. [Google Scholar] [PubMed]
- Wu, Q.; Wei, Z.; Yang, Z.; Wang, T.; Ren, L.; Hu, X.; Meng, Q.; Guo, Y.; Zhu, Q.; Robert, J.; et al. Phylogeny, genomic organization and expression of lambda and kappa immunoglobulin light chain genes in a reptile, Anolis carolinensis. Dev. Comp. Immunol. 2010, 34, 579–589. [Google Scholar] [PubMed]
- Work, T.M.; Balazs, G.H.; Rameyer, R.A.; Chang, S.P.; Berestecky, J. Assessing humoral and cell-mediated immune response in Hawaiian green turtles, Chelonia mydas. Vet. Immunol. Immunopathol. 2000, 74, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Grey, H.M. Phylogeny of the immune response. Studies on some physical chemical and serologic characteristics of antibody produced in the turtle. J. Immunol. 1963, 91, 819–825. [Google Scholar] [PubMed]
- Cooper, E.L.; Klempau, A.E.; Zapata, A.G. Introduction to reptilian immunity. In Biology of the Reptilia; Gans, C., Billett, F., Maderson, P.F.A., Eds.; John Wiley & Sons: New York, NY, USA, 1985; Volume 14, pp. 601–678. [Google Scholar]
- Marschang, R.E.; Gravendyck, M.; Kaleta, E.F. Herpesviruses in tortoises: Investigations into virus isolation and the treatment of viral stomatitis in Testudo hermanni and T. graeca. Zentralbl Veterinarmed B 1997, 44, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Merchant, M.E.; Roche, C.; Elsey, R.M.; Prudhomme, J. Antibacterial properties of serum from the American alligator (Alligator mississippiensis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003, 136, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Pasmans, F.; Herdt, P.D.; Nerom, A.V.; Haesebrouck, F. Induction of the respiratory burst in turtle peritoneal macrophages by Salmonella muenchen. Dev. Comp. Immunol. 2001, 25, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Cone, R.E.; Marchalonis, J.J. Cellular and humoral aspects of the influence of environmental temperature on the immune response of poikilothermic vertebrates. J. Immunol. 1972, 108, 952–957. [Google Scholar] [PubMed]
- Zapata, A.G.; Varas, A.; Torroba, M. Seasonal variations in the immune system of lower vertebrates. Immunol. Today 1992, 13, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F. Re-evaluation of the immunological Big Bang. Curr. Biol. 2014, 24, R1060–R1065. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.W. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pettinello, R.; Dooley, H. The Immunoglobulins of Cold-Blooded Vertebrates. Biomolecules 2014, 4, 1045-1069. https://doi.org/10.3390/biom4041045
Pettinello R, Dooley H. The Immunoglobulins of Cold-Blooded Vertebrates. Biomolecules. 2014; 4(4):1045-1069. https://doi.org/10.3390/biom4041045
Chicago/Turabian StylePettinello, Rita, and Helen Dooley. 2014. "The Immunoglobulins of Cold-Blooded Vertebrates" Biomolecules 4, no. 4: 1045-1069. https://doi.org/10.3390/biom4041045