Functional Integration of mRNA Translational Control Programs
Abstract
:1. Regulated mRNA Translation and Control of Cell Cycle



2. Multiple RBP Input Is a Prevalent Regulatory Mechanism for Control of mRNA Translation
| HOMO SAPIENS (66,974 Total 3' UTRs) | MUS MUSCULUS (26,420 Total 3' UTRs) | XENOPUS (26,511 Total 3' UTRs) | |||
|---|---|---|---|---|---|
| Sequence Motif Combinations | Matching 3' UTRs | Sequence Motif Combinations | Matching 3' UTRs | Sequence Motif Combinations | Matching 3' UTRs |
| PBE | 15,156 | PBE | 5444 | PBE | 6312 |
| & MBE | 14,437 | & MBE | 5136 | & MBE | 5521 |
| NOT MBE | 719 | NOT MBE | 308 | NOT MBE | 791 |
| & CPE | 13,536 | & CPE | 4693 | & CPE | 5427 |
| NOT CPE | 1620 | NOT CPE | 751 | NOT CPE | 885 |
| & TCS | 9462 | & TCS | 3101 | & TCS | 2738 |
| NOT TCS | 5694 | NOT TCS | 2343 | NOT TCS | 3574 |
| NOT (MBE OR CPE) | 409 | NOT (MBE OR CPE) | 174 | NOT (MBE OR CPE) | 314 |
| NOT (MBE OR TCS) | 613 | NOT (MBE OR TCS) | 272 | NOT (MBE OR TCS) | 638 |
| NOT (CPE OR TCS) | 1287 | NOT (CPE OR TCS) | 606 | NOT (CPE OR TCS) | 735 |
| NOT (MBE OR CPE OR TCS) | 373 | NOT (MBE OR CPE OR TCS) | 162 | NOT (MBE OR CPE OR TCS) | 273 |
| MBE | 50,101 | MBE | 20,079 | MBE | 18,912 |
| & PBE | 14,437 | & PBE | 5136 | & PBE | 5521 |
| NOT PBE | 35,664 | NOT PBE | 14,943 | NOT PBE | 13,391 |
| & CPE | 37,483 | & CPE | 14,215 | & CPE | 15,101 |
| NOT CPE | 12,618 | NOT CPE | 5864 | NOT CPE | 3811 |
| & TCS | 21,858 | & TCS | 8358 | & TCS | 6861 |
| NOT TCS | 28,243 | NOT TCS | 11,721 | NOT TCS | 12,051 |
| NOT (PBE OR CPE) | 11,407 | NOT (PBE OR CPE) | 5287 | NOT (PBE OR CPE) | 3240 |
| NOT (PBE OR TCS) | 23,162 | NOT (PBE OR TCS) | 9650 | NOT (PBE OR TCS) | 9115 |
| NOT (CPE OR TCS) | 10,670 | NOT (CPE OR TCS) | 4790 | NOT (CPE OR TCS) | 3233 |
| NOT (PBE OR CPE OR TCS) | 9756 | NOT (PBE OR CPE OR TCS) | 4346 | NOT (PBE OR CPE OR TCS) | 2771 |
| CPE | 41,567 | CPE | 15,698 | CPE | 18,383 |
| & PBE | 13,536 | & PBE | 4693 | & PBE | 5427 |
| NOT PBE | 28,031 | NOT PBE | 11,005 | NOT PBE | 12,956 |
| & MBE | 37,483 | & MBE | 14,215 | & MBE | 15,101 |
| NOT MBE | 4084 | NOT MBE | 1483 | NOT MBE | 3282 |
| & TCS | 20,638 | & TCS | 7518 | & TCS | 6926 |
| NOT TCS | 20,929 | NOT TCS | 8180 | NOT TCS | 11,457 |
| NOT (PBE OR MBE) | 3774 | NOT (PBE OR MBE) | 1349 | NOT (PBE OR MBE) | 2805 |
| NOT (PBE OR TCS) | 16,522 | NOT (PBE OR TCS) | 6443 | NOT (PBE OR TCS) | 8618 |
| NOT (MBE OR TCS) | 3356 | NOT (MBE OR TCS) | 1249 | NOT (MBE OR TCS) | 2639 |
| NOT (PBE OR MBE OR TCS) | 3116 | NOT (PBE OR MBE OR TCS) | 1139 | NOT (PBE OR MBE OR TCS) | 2274 |
| TCS | 23,235 | TCS | 8880 | TCS | 7827 |
| & PBE | 9462 | & PBE | 3101 | & PBE | 2738 |
| NOT PBE | 13,773 | NOT PBE | 5779 | NOT PBE | 5089 |
| & MBE | 21,858 | & MBE | 8358 | & MBE | 6861 |
| NOT MBE | 1377 | NOT MBE | 522 | NOT MBE | 966 |
| & CPE | 20,638 | & CPE | 7518 | & CPE | 6926 |
| NOT CPE | 2597 | NOT CPE | 1362 | NOT CPE | 901 |
| NOT (PBE OR MBE) | 1271 | NOT (PBE OR MBE) | 486 | NOT (PBE OR MBE) | 813 |
| NOT (PBE OR CPE) | 2264 | NOT (PBE OR CPE) | 1217 | NOT (PBE OR CPE) | 751 |
| NOT (MBE OR CPE) | 649 | NOT (MBE OR CPE) | 288 | NOT (MBE OR CPE) | 323 |
| NOT (PBE OR MBE OR CPE) | 613 | NOT (PBE OR MBE OR CPE) | 276 | NOT (PBE OR MBE OR CPE) | 282 |
| NO MATCH | 11,767 | NO MATCH | 4408 | NO MATCH | 3721 |
3. Advantages of mRNA Combinatorial Control
4. RBP-Specific Co-Factor Interactions
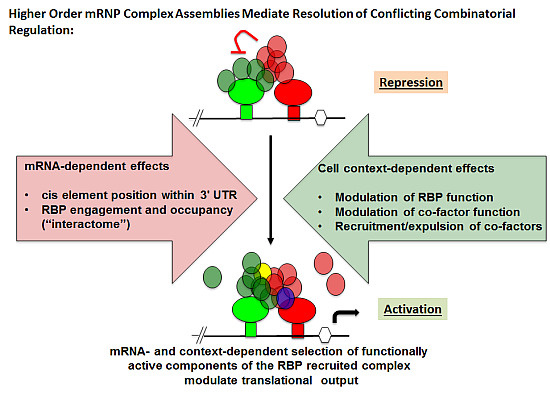
5. Experimental Evidence for Adaptive Regulatory Assemblies in the Control of Translation


| pGBK DNA Binding Fusion Vector | pACT Transcriptional Activation Fusion Vector | Interaction? |
|---|---|---|
| Empty | Musashi1 | No |
| Musashi1 | Empty | No |
| Musashi1 | CPEB1 | No |
| Musashi1 | CPEB4 | No |
| Musashi1 | Musashi1 | Yes |
6. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blackinton, J.G.; Keene, J.D. Post-Transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell. Dev. Biol. 2014, 34, 44–54. [Google Scholar] [CrossRef] [PubMed]
- D'Ambrogio, A.; Nagaoka, K.; Richter, J.D. Translational control of cell growth and malignancy by the cpebs. Nat. Rev. 2013. [Google Scholar] [CrossRef] [PubMed]
- De Andres-Aguayo, L.; Varas, F.; Graf, T. Musashi 2 in hematopoiesis. Curr. Opin. Hematol. 2012, 19, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Larsson, O.; Tian, B.; Sonenberg, N. Toward a genome-wide landscape of translational control. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, X. Concise review: Fragile x proteins in stem cell maintenance and differentiation. Stem Cells 2014, 32, 1724–1733. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Daley, G.Q. Lin28: Primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013, 12, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, C.R.; Moreno, M.V.; Olshen, A.B.; Taylor, B.S.; Ruggero, D. The translational landscape of the mammalian cell cycle. Mol. Cell 2013, 52, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.E.; Ciosk, R. RNA-Based regulation of pluripotency. Trends Genet. 2013, 29, 99–107. [Google Scholar] [CrossRef] [PubMed]
- MacNicol, A.M.; Wilczynska, A.; MacNicol, M.C. Function and regulation of the mammalian musashi mRNA translational regulator. Biochem. Soc. Trans. 2008, 36, 528–530. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.A. A cancer fate in the hands of a samurai. Nat. Med. 2010, 16, 963–965. [Google Scholar] [CrossRef] [PubMed]
- Okano, H.; Kawahara, H.; Toriya, M.; Nakao, K.; Shibata, S.; Imai, T. Function of RNA-binding protein musashi-1 in stem cells. Exp. Cell Res. 2005, 306, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Ruohola-Baker, H. Regulation of stem cell populations by microRNAs. Adv. Exp. Med. Biol. 2013, 786, 329–351. [Google Scholar] [PubMed]
- Alison, M.R.; Lim, S.M.; Nicholson, L.J. Cancer stem cells: Problems for therapy? J. Pathol. 2011, 223, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.J.; Graham, T.A. Cancer: Resolving the stem-cell debate. Nature 2012, 488, 462–463. [Google Scholar] [CrossRef] [PubMed]
- Dontu, G.; Al-Hajj, M.; Abdallah, W.M.; Clarke, M.F.; Wicha, M.S. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003, 36, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Pece, S.; Tosoni, D.; Confalonieri, S.; Mazzarol, G.; Vecchi, M.; Ronzoni, S.; Bernard, L.; Viale, G.; Pelicci, P.G.; di Fiore, P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010, 140, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.J.; Fleming, J.M.; Ali, M.A.; Pesesky, M.W.; Ginsburg, E.; Vonderhaar, B.K. Dynamic regulation of CD24 and the invasive, CD44posCD24neg phenotype in breast cancer cell lines. Breast Cancer Res. 2009. [Google Scholar] [CrossRef] [PubMed]
- Goffart, N.; Kroonen, J.; Rogister, B. Glioblastoma-Initiating cells: Relationship with neural stem cells and the micro-environment. Cancers 2013, 5, 1049–1071. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Yang, K.; Rich, J.N. The evolving landscape of glioblastoma stem cells. Curr. Opin. Neurol. 2013, 26, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Ortensi, B.; Setti, M.; Osti, D.; Pelicci, G. Cancer stem cell contribution to glioblastoma invasiveness. Stem Cell Res. Ther. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Sureban, S.M.; May, R.; George, R.J.; Dieckgraefe, B.K.; McLeod, H.L.; Ramalingam, S.; Bishnupuri, K.S.; Natarajan, G.; Anant, S.; Houchen, C.W. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology 2008, 134, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Penalva, L.O.; Yuan, H.; Linnoila, R.I.; Lu, J.; Okano, H.; Glazer, R.I. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol. Cancer 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Yu, H.; Linnoila, R.I.; Li, L.; Li, D.; Mo, B.; Okano, H.; Penalva, L.O.; Glazer, R.I. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget 2013, 4, 739–750. [Google Scholar] [PubMed]
- Grzmil, M.; Hemmings, B.A. Translation regulation as a therapeutic target in cancer. Cancer Res. 2012, 72, 3891–3900. [Google Scholar] [CrossRef] [PubMed]
- Kuersten, S.; Radek, A.; Vogel, C.; Penalva, L.O. Translation regulation gets its “omics” moment. Wiley Interdiscip. Rev. RNA 2013, 4, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.R. Ribosomal footprints on a transcriptome landscape. Genome Biol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, F.; Preiss, T.; Hentze, M.W. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb. Perspect. Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.C.; Ephrussi, A. mRNA localization: Gene expression in the spatial dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Pratt, G.; Yeo, G.W.; Moore, M.J. The clothes make the mRNA: Past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015, 84, 325–354. [Google Scholar] [CrossRef] [PubMed]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, K.; Wang, Y.; Hardy, L.L.; MacNicol, M.C.; MacNicol, A.M. Enforcing temporal control of maternal mRNA translation during oocyte cell cycle progression. EMBO J. 2010, 29, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, A.; Cox, L.L.; MacNicol, A.M. Cytoplasmic polyadenylation element (cpe)- and cpe-binding protein (cpeb)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 2004, 279, 17650–17659. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, A.; Wilczynska, A.; Thampi, P.; Cox, L.L.; MacNicol, A.M. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006, 25, 2792–2801. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Deering, R.P.; Lu, Y.; Tivnan, P.; Lianoglou, S.; Al-Shahrour, F.; Ebert, B.L.; Hacohen, N.; Leslie, C.; Daley, G.Q.; et al. Musashi-2 controls cell fate, lineage bias, and TGF-beta signaling in hscs. J. Exp. Med. 2014, 211, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, C.E.; Lau, H.T.; Mangan, H.; Hardy, L.L.; Sunnotel, O.; Guo, F.; MacNicol, A.M.; Walsh, C.P.; Lees-Murdock, D.J. Efficient translation of dnmt1 requires cytoplasmic polyadenylation and musashi binding elements. PLoS ONE 2014, 9, e88385. [Google Scholar] [CrossRef] [PubMed]
- Cragle, C.E.; MacNicol, A.M. From oocyte to fertilizable egg—Regulated mRNA translation and the control of maternal gene expression. In Xenopus Development; Kloc, M., Kubiak, J.Z., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar] [CrossRef]
- MacNicol, M.C.; MacNicol, A.M. Developmental timing of mRNA translation—Integration of distinct regulatory elements. Mol. Reprod. Dev. 2010, 77, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Padmanabhan, K.; Richter, J.D. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA 2010, 16, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, K.; Richter, J.D. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 2006, 20, 199–209. [Google Scholar] [CrossRef] [PubMed]
- de Moor, C.H.; Richter, J.D. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999, 18, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Novoa, I.; Gallego, J.; Ferreira, P.G.; Mendez, R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat. Cell Biol. 2010, 12, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, A.; Yamamoto, T.M.; Cook, J.M.; Silva, K.D.; Kotter, C.V.; Carter, G.S.; Holt, J.W.; Lavender, H.F.; MacNicol, A.M.; Ying Wang, Y.; et al. Xenopus laevis zygote arrest 2 (zar2) encodes a zinc finger RNA-binding protein that binds to the translational control sequence in the maternal Wee1 mRNA and regulates translation. Dev. Biol. 2012, 369, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.M.; Cook, J.M.; Kotter, C.V.; Khat, T.; Silva, K.D.; Ferreyros, M.; Holt, J.W.; Knight, J.D.; Charlesworth, A. Zar1 represses translation in Xenopus oocytes and binds to the TCS in maternal mRNAs with different characteristics than zar2. Biochim. Biophys. Acta 2013, 1829, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Ballantyne, S.; Daniel, D.L.J.; Wickens, M. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell 1997, 8, 1633–1648. [Google Scholar] [CrossRef] [PubMed]
- De Moor, C.H.; Richter, J.D. The mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell Biol. 1997, 17, 6419–6426. [Google Scholar] [PubMed]
- Ivshina, M.; Lasko, P.; Richter, J.D. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 2014, 30, 393–415. [Google Scholar] [CrossRef] [PubMed]
- MacNicol, M.C.; Cragle, C.E.; MacNicol, A.M. Context-dependent regulation of musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle 2011, 10, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, K.; MacNicol, M.C.; Wang, Y.; Cragle, C.E.; Tackett, A.J.; Hardy, L.L.; MacNicol, A.M. Ringo/CDK and map kinase regulate the activity of the cell fate determinant musashi to promote cell cycle re-entry in Xenopus oocytes. J. Biol. Chem. 2012, 287, 10639–10649. [Google Scholar] [CrossRef] [PubMed]
- Keady, B.T.; Kuo, P.; Martinez, S.E.; Yuan, L.; Hake, L.E. Mapk interacts with XGeF and is required for CPEB activation during meiosis in Xenopus oocytes. J. Cell Sci. 2007, 120, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Igea, A.; Mendez, R. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 2010, 29, 2182–2193. [Google Scholar] [CrossRef] [PubMed]
- Reverte, C.G.; Ahearn, M.D.; Hake, L.E. Cpeb degradation during xenopus oocyte maturation requires a pest domain and the 26s proteasome. Dev. Biol. 2001, 231, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Mendez, R.; Barnard, D.; Richter, J.D. Differential mRNA translation and meiotic progression require CDC2-mediated CPEB destruction. EMBO J. 2002, 21, 1833–1844. [Google Scholar] [CrossRef] [PubMed]
- Nechama, M.; Lin, C.L.; Richter, J.D. An unusual two-step control of CPEB destruction by PIN1. Mol. Cell Biol. 2013, 33, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Grillo, G.; Turi, A.; Licciulli, F.; Mignone, F.; Liuni, S.; Banfi, S.; Gennarino, V.A.; Horner, D.S.; Pavesi, G.; Picardi, E.; et al. Utrdb and utrsite (release 2010): A collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010, 38, D75–D80. [Google Scholar] [CrossRef] [PubMed]
- De Moor, C.H.; Meijer, H.; Lissenden, S. Mechanisms of translational control by the 3' UTR in development and differentiation. Semin. Cell Dev. Biol. 2005, 16, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Grillo, G.; Licciulli, F.; Liuni, S.; Sbisa, E.; Pesole, G. Patsearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003, 31, 3608–3612. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, S.; Kotani, T.; Mita, K.; Kawasaki, T.; Katsu, Y.; Nagahama, Y.; Yamashita, M. Involvement of xenopus pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 2003, 120, 865–880. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Charlesworth, A.; Byrd, S.M.; Gregerson, R.; MacNicol, M.C.; MacNicol, A.M. A novel mRNA 3' untranslated region translational control sequence regulates Xenopus Wee1 mRNA translation. Dev. Biol. 2008, 317, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, A.; Ridge, J.A.; King, L.A.; MacNicol, M.C.; MacNicol, A.M. A novel regulatory element determines the timing of mos mRNA translation during Xenopus oocyte maturation. EMBO J. 2002, 21, 2798–2806. [Google Scholar] [CrossRef] [PubMed]
- Pique, M.; Lopez, J.M.; Foissac, S.; Guigo, R.; Mendez, R. A combinatorial code for CPE-mediated translational control. Cell 2008, 132, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.N.; Habermacher, R.; Martine, U.; Closs, E.I.; Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006, 125, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Kundu, P.; Fabian, M.R.; Sonenberg, N.; Bhattacharyya, S.N.; Filipowicz, W. Hur protein attenuates miRNA-mediated repression by promoting mirisc dissociation from the target RNA. Nucleic Acids Res. 2012, 40, 5088–5100. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Mazan-Mamczarz, K.; Kawai, T.; Yang, X.; Martindale, J.L.; Gorospe, M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004, 23, 3092–3102. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, A.; Welk, J.; MacNicol, A. The temporal control of wee1 mRNA translation during Xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3' untranslated region. Dev. Biol. 2000, 227, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Radford, H.E.; Meijer, H.A.; de Moor, C.H. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta 2008, 1779, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Stebbins-Boaz, B.; Cao, Q.; de Moor, C.; Mendez, R.; Richter, J.D. Maskin is a CPEB-associated factor that transiently interacts with eIF-4E. Mol. Cell 1999, 4, 1017–1027. [Google Scholar] [CrossRef]
- Minshall, N.; Reiter, M.H.; Weil, D.; Standart, N. Cpeb interacts with an ovary-specific eIF4E and 4E-T in early xenopus oocytes. J. Biol. Chem. 2007, 282, 37389–37401. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Richter, J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 2006, 24, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Barnard, D.C.; Cao, Q.; Richter, J.D. Differential phosphorylation controls maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol. Cell Biol. 2005, 25, 7605–7615. [Google Scholar] [CrossRef] [PubMed]
- Cragle, C.; Macnicol, A.M. Musashi-directed translational activation of target mRNAs is mediated by the poly[A] polymerase, germline development-2. J. Biol. Chem. 2014, 289, 14239–14251. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Imai, T.; Imataka, H.; Tsujimoto, M.; Matsumoto, K.; Okano, H. Neural RNA-binding protein musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008, 181, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.W.; Blee, T.K.; Gray, N.K. Poly(A)-binding proteins are required for diverse biological processes in metazoans. Biochem. Soc. Trans. 2014, 42, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- MacNicol, M.C.; Hardy, L.L.; Spencer, H.J.; MacNicol, A.M. Neural stem and progenitor cell fate transition requires regulation of musashi1 function. BMC Dev. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Nagata, T.; Tsuda, K.; Kobayashi, N.; Imai, T.; Okano, H.; Yamazaki, T.; Katahira, M. Structure of musashi1 in a complex with target RNA: The role of aromatic stacking interactions. Nucleic Acids Res. 2012, 40, 3218–3231. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Okada, Y.; Imai, T.; Iwanami, A.; Mischel, P.S.; Okano, H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 2011, 286, 16121–16130. [Google Scholar] [CrossRef] [PubMed]
- Wuebben, E.L.; Mallanna, S.K.; Cox, J.L.; Rizzino, A. Musashi2 is required for the self-renewal and pluripotency of embryonic stem cells. PLoS ONE 2012, 7, e34827. [Google Scholar] [CrossRef] [PubMed]
- Szabat, M.; Kalynyak, T.B.; Lim, G.E.; Chu, K.Y.; Yang, Y.H.; Asadi, A.; Gage, B.K.; Ao, Z.; Warnock, G.L.; Piret, J.M.; et al. Musashi expression in beta-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2011, 2, e232. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Kwon, H.Y.; Zimdahl, B.; Congdon, K.L.; Blum, J.; Lento, W.E.; Zhao, C.; Lagoo, A.; Gerrard, G.; Foroni, L.; et al. Regulation of myeloid leukaemia by the cell-fate determinant musashi. Nature 2010, 466, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Kharas, M.G.; Lengner, C.J.; Al-Shahrour, F.; Bullinger, L.; Ball, B.; Zaidi, S.; Morgan, K.; Tam, W.; Paktinat, M.; Okabe, R.; et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010, 16, 903–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Richter, J.D. Ringo/CDK1 and CPEB mediate poly(A) tail stabilization and translational regulation by EPAB. Genes Dev. 2007, 21, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Voeltz, G.K.; Ongkasuwan, J.; Standart, N.; Steitz, J.A. A novel embryonic poly(A) binding protein, EPAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001, 15, 774–788. [Google Scholar] [CrossRef] [PubMed]
- MacNicol, M.C.; Muslin, A.J.; MacNicol, A.M. Disruption of the 14-3-3 binding site within the B-Raf kinase domain uncouples catalytic activity from PC12 cell differentiation. J. Biol. Chem. 2000, 275, 3803–3809. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Huang, Y.T.; Richter, J.D. Transient CPEB dimerization and translational control. RNA 2012, 18, 1050–1061. [Google Scholar] [CrossRef] [PubMed]
- Schelhorn, C.; Gordon, J.M.; Ruiz, L.; Alguacil, J.; Pedroso, E.; Macias, M.J. RNA recognition and self-association of CPEB4 is mediated by its tandem RRM domains. Nucleic Acids Res. 2014, 42, 10185–10195. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Welk, J.; Charlesworth, A.; Smith, G.D.; MacNicol, A.M. Identification of the gene encoding human cytoplasmic polyadenylation element binding protein. Gene 2001, 263, 113–121. [Google Scholar] [CrossRef]
- MacNicol, M.C.; Pot, D.; MacNicol, A.M. pXen, a utility vector for the expression of GST-fusion proteins in Xenopus laevis oocytes and embryos. Gene 1997, 196, 25–29. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
MacNicol, M.C.; Cragle, C.E.; Arumugam, K.; Fosso, B.; Pesole, G.; MacNicol, A.M. Functional Integration of mRNA Translational Control Programs. Biomolecules 2015, 5, 1580-1599. https://doi.org/10.3390/biom5031580
MacNicol MC, Cragle CE, Arumugam K, Fosso B, Pesole G, MacNicol AM. Functional Integration of mRNA Translational Control Programs. Biomolecules. 2015; 5(3):1580-1599. https://doi.org/10.3390/biom5031580
Chicago/Turabian StyleMacNicol, Melanie C., Chad E. Cragle, Karthik Arumugam, Bruno Fosso, Graziano Pesole, and Angus M. MacNicol. 2015. "Functional Integration of mRNA Translational Control Programs" Biomolecules 5, no. 3: 1580-1599. https://doi.org/10.3390/biom5031580