Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function
Abstract
:1. Introduction
1.1. Post-Transcriptional Regulation
1.2. mRNA Stability in Neurons
1.3. Mechanisms Controlling mRNA Stability
2. RNA-Binding Proteins
2.1. HuD and Other Members of ELAV-Like/Hu Protein Family
2.2. KSRP
2.3. ZBP1/IMP1
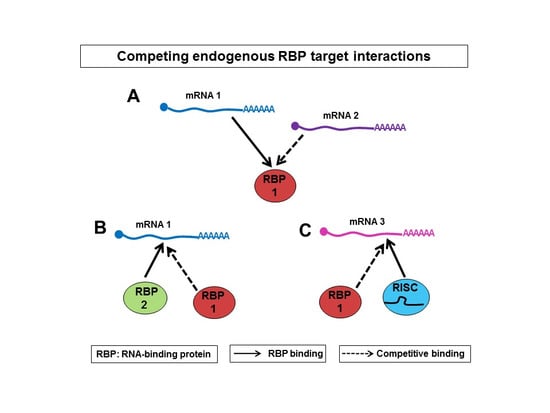
3. Competition between mRNAs for Binding to RBPs

4. Competition vs. Cooperation of RBPs
5. Competition vs. Cooperation between RBPs and miRNAs
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wilusz, C.J.; Wormington, M.; Peltz, S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001, 2, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Brewer, G. The regulation of mRNA stability in mammalian cells: 2.0. Gene 2012, 500, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Ross, J. mRNA stability in mammalian cells. Microbiol. Rev. 1995, 59, 423–450. [Google Scholar] [PubMed]
- Shaw, G.; Kamen, R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 1986, 46, 659–667. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef]
- Bakheet, T.; Frevel, M.; Williams, B.R.; Greer, W.; Khabar, K.S. ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001, 29, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Frevel, M.A.; Bakheet, T.; Silva, A.M.; Hissong, J.G.; Khabar, K.S.; Williams, B.R. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell Biol. 2003, 23, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Contente-Cuomo, T.; Perrone-Bizzozero, N.I. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010, 38, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Perrone-Bizzozero, N.I. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci. Res. 2008, 86, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Tenenbaum, S.A.; Carson, C.C.; Lager, P.J.; Keene, J.D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 2000, 97, 14085–14090. [Google Scholar] [CrossRef] [PubMed]
- Tiruchinapalli, D.M.; Ehlers, M.D.; Keene, J.D. Activity-dependent expression of RNA binding protein HuD and its association with mRNAs in neurons. RNA Biol. 2008, 5, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Westmark, C.J.; Gourronc, F.A.; Bartleson, V.B.; Sayin, U.; Bhattacharya, S.; Sutula, T.; Malter, J.S. HuR mRNA ligands expressed after seizure. J. Neuropathol. Exp. Neurol. 2005, 64, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Shyu, A.B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2011, 2, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Petfalski, E.; Shevchenko, A.; Mann, M.; Tollervey, D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3'→5' exoribonucleases. Cell 1997, 91, 457–466. [Google Scholar] [CrossRef]
- Van Hoof, A.; Parker, R. The exosome: A proteasome for RNA? Cell 1999, 99, 347–350. [Google Scholar] [CrossRef]
- Chen, C.Y.; Gherzi, R.; Ong, S.E.; Chan, E.L.; Raijmakers, R.; Pruijn, G.J.; Stoecklin, G.; Moroni, C.; Mann, M.; Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 2001, 107, 451–464. [Google Scholar] [CrossRef]
- Mukherjee, D.; Gao, M.; O’Connor, J.P.; Raijmakers, R.; Pruijn, G.; Lutz, C.S.; Wilusz, J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002, 21, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin, G.; Mayo, T.; Anderson, P. ARE-mRNA degradation requires the 5'–3' decay pathway. EMBO Rep. 2006, 7, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Fenger-Gron, M.; Fillman, C.; Norrild, B.; Lykke-Andersen, J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 2005, 20, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, C.J.; Wilusz, J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004, 20, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Linker, K.; Pautz, A.; Fechir, M.; Hubrich, T.; Greeve, J.; Kleinert, H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005, 33, 4813–4827. [Google Scholar] [CrossRef] [PubMed]
- Gherzi, R.; Chen, C.Y.; Trabucchi, M.; Ramos, A.; Briata, P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip. Rev. RNA 2010, 1, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Briata, P.; Chen, C.Y.; Ramos, A.; Gherzi, R. Functional and molecular insights into KSRP function in mRNA decay. Biochim. Biophys. Acta 2013, 1829, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Briata, P.; Ilengo, C.; Corte, G.; Moroni, C.; Rosenfeld, M.G.; Chen, C.Y.; Gherzi, R. The Wnt/beta-catenin→Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol. Cell 2003, 12, 1201–1211. [Google Scholar] [CrossRef]
- Briata, P.; Forcales, S.V.; Ponassi, M.; Corte, G.; Chen, C.Y.; Karin, M.; Puri, P.L.; Gherzi, R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 2005, 20, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Amirouche, A.; Tadesse, H.; Lunde, J.A.; Belanger, G.; Cote, J.; Jasmin, B.J. Activation of p38 signaling increases utrophin A expression in skeletal muscle via the RNA-binding protein KSRP and inhibition of AU-rich element-mediated mRNA decay: Implications for novel DMD therapeutics. Hum. Mol. Genet. 2013, 22, 3093–3111. [Google Scholar] [CrossRef] [PubMed]
- Dalmau, J.; Furneaux, H.M.; Gralla, R.J.; Kris, M.G.; Posner, J.B. Detection of the anti-Hu antibody in the serum of patients with small cell lung cancer—A quantitative western blot analysis. Ann. Neurol. 1990, 27, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Robinow, S.; Campos, A.R.; Yao, K.M.; White, K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science 1988, 242, 1570–1572. [Google Scholar] [CrossRef] [PubMed]
- Okano, H.J.; Darnell, R.B. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 1997, 17, 3024–3037. [Google Scholar] [PubMed]
- Kenan, D.J.; Query, C.C.; Keene, J.D. RNA recognition: Towards identifying determinants of specificity. Trends Biochem. Sci. 1991, 16, 214–220. [Google Scholar] [CrossRef]
- Keene, J.D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA 1999, 96, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Marusich, M.F.; Furneaux, H.M.; Henion, P.D.; Weston, J.A. Hu neuronal proteins are expressed in proliferating neurogenic cells. J. Neurobiol. 1994, 25, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Wakamatsu, Y.; Weston, J.A. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development 1997, 124, 3449–3460. [Google Scholar] [PubMed]
- Peng, S.S.-Y.; Chen, C.-Y.A.; Xu, N.; Shyu, A.-B. RNA stabilization of the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998, 17, 3461–3470. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.C.; Steitz, J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. PNAS 1998, 95, 15293–15298. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.N.; Zhou, H.L.; Sharma, A.; Lou, H. All three RNA recognition motifs and the hinge region of HuC play distinct roles in the regulation of alternative splicing. Nucleic Acids Res. 2013, 41, 5049–5061. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.D.; Sengupta, J.; Morin, M.; Neve, R.L.; Valenzuela, C.F.; Perrone-Bizzozero, N.I. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp. Neurol. 2001, 168, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Merhege, M.A.; Twiss, J.; Perrone-Bizzozero, N.I. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: Association with polysomes and upregulation during contextual learning. Neurosci. Lett. 2004, 371, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Tanner, D.C.; Merhege, M.; Deschenes-Furry, J.; Jasmin, B.; Perrone-Bizzozero, N.I. In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD. J. Neurochem. 2006, 96, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Kasashima, K.; Terashima, K.; Yamamoto, K.; Sakashita, E.; Sakamoto, H. Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells 1999, 4, 667–683. [Google Scholar] [CrossRef] [PubMed]
- Clayton, G.H.; Perez, G.M.; Smith, R.L.; Owens, G.C. Expression of mRNA for the ELAV-like neural-specific RNA binding protein, HuD, during nervous system development. Dev. Brain Res. 1998, 109, 271–280. [Google Scholar] [CrossRef]
- Akamatsu, W.; Okano, H.J.; Osumi, N.; Inoue, T.; Nakamura, S.; Sakakibara, S.; Miura, M.; Matsuo, N.; Darnell, R.B.; Okano, H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc. Natl. Acad. Sci. USA 1999, 96, 9885–9890. [Google Scholar] [CrossRef] [PubMed]
- Mobarak, C.D.; Anderson, K.D.; Morin, M.; Beckel-Mitchener, A.; Rogers, S.L.; Furneaux, H.; King, P.; Perrone-Bizzozero, N.I. The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol. Biol. Cell 2000, 11, 3191–3203. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.D.; Morin, M.A.; Beckel-Mitchener, A.; Mobarak, C.D.; Neve, R.L.; Furneaux, H.M.; Burry, R.; Perrone-Bizzozero, N.I. Overexpression of HuD, but not of its truncated form HuD I + II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J. Neurochem. 2000, 75, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Dobashi, Y.; Shoji, M.; Wakata, Y.; Kameya, T. Expression of HuD protein is essential for initial phase of neuronal differentiation in rat pheochromocytoma cells. Biochem. Biophys. Res. Comm. 1998, 244, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, W.; Fujihara, H.; Mitsuhashi, T.; Yano, M.; Shibata, S.; Hayakawa, Y.; Okano, H.J.; Sakakibara, S.; Takano, H.; Takano, T.; et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl. Acad. Sci. USA 2005, 102, 4625–4630. [Google Scholar] [CrossRef] [PubMed]
- DeBoer, E.M.; Azevedo, R.; Vega, T.A.; Brodkin, J.; Akamatsu, W.; Okano, H.; Wagner, G.C.; Rasin, M.R. Prenatal deletion of the RNA-binding protein HuD disrupts postnatal cortical circuit maturation and behavior. J. Neurosci. 2014, 34, 3674–3686. [Google Scholar] [CrossRef] [PubMed]
- Ince-Dunn, G.; Okano, H.J.; Jensen, K.B.; Park, W.Y.; Zhong, R.; Ule, J.; Mele, A.; Fak, J.J.; Yang, C.; Zhang, C.; et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 2012, 75, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Quattrone, A.; Pascale, A.; Nogues, X.; Zhao, W.; Gusev, P.; Pacini, A.; Alkon, D.L. Posttranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 11668–11673. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Gusev, P.A.; Amadio, M.; Dottorini, T.; Govoni, S.; Alkon, D.L.; Quattrone, A. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc. Natl. Acad. Sci. USA 2004, 101, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Tanner, D.C.; Nixon, S.; Okano, H.J.; Okano, H.; Perrone-Bizzozero, N.I. Coordinated expression of HuD and GAP-43 in hippocampal dentate granule cells during developmental and adult plasticity. Neurochem. Res. 2007, 32, 2142–2151. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Qiu, S.; Tanner, D.C.; Paik, J.; Perrone-Bizzozero, N.I.; Weeber, E.J. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol. Learn. Mem. 2007, 87, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Fukao, A.; Sasano, Y.; Imataka, H.; Inoue, K.; Sakamoto, H.; Sonenberg, N.; Thoma, C.; Fujiwara, T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol. Cell 2009, 36, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Fukao, A.; Mishima, Y.; Takizawa, N.; Oka, S.; Imataka, H.; Pelletier, J.; Sonenberg, N.; Thoma, C.; Fujiwara, T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol. Cell 2014, 56, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Beckel-Mitchener, A.C.; Miera, A.; Keller, R.; Perrone-Bizzozero, N.I. Poly (A) tail length dependent stabilization of GAP-43 mRNA by the RNA binding protein HuD. J. Biol. Chem. 2002, 28, 27996–28002. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Kim, H.H.; Kim, P.; Donnelly, C.J.; Kalinski, A.L.; Vuppalanchi, D.; Park, M.; Lee, S.J.; Merianda, T.T.; Perrone-Bizzozero, N.I.; et al. A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3' untranslated region AU-rich regulatory element. J. Neurochem. 2013, 126, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.; Behar, L.; Elliott, E.; Ginzburg, I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 2004, 89, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Akten, B.; Kye, M.J.; Haole, T.; Wertz, M.H.; Singh, S.; Nie, D.; Huang, J.; Merianda, T.T.; Twiss, J.L.; Beattie, C.E.; et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. USA 2011, 108, 10337–10342. [Google Scholar] [CrossRef] [PubMed]
- Sosanya, N.M.; Huang, P.P.; Cacheaux, L.P.; Chen, C.J.; Nguyen, K.; Perrone-Bizzozero, N.I.; Raab-Graham, K.F. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J. Cell Biol. 2013, 202, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Sosanya, N.M.; Cacheaux, L.P.; Workman, E.R.; Niere, F.; Perrone-Bizzozero, N.I.; Raab-Graham, K.F. Mammalian target of rapamycin (mTOR) tagging promotes dendritic branch variability through the Capture of Ca2+/Calmodulin-dependent protein kinase II alpha (CaMKIIalpha) mRNAs by the RNA-binding protein HuD. J. Biol. Chem. 2015, 290, 16357–16371. [Google Scholar] [CrossRef] [PubMed]
- Vanevski, F.; Xu, B. HuD interacts with BDNF mRNA and is essential for activity-induced BDNF synthesis in dendrites. PLoS ONE 2015, 10, e0117264. [Google Scholar] [CrossRef] [PubMed]
- Min, H.; Turck, C.W.; Nikolic, J.M.; Black, D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997, 11, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Gherzi, R.; Lee, K.Y.; Briata, P.; Wegmuller, D.; Moroni, C.; Karin, M.; Chen, C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 2004, 14, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Snee, M.; Kidd, G.J.; Munro, T.P.; Smith, R. RNA trafficking and stabilization elements associate with multiple brain proteins. J. Cell Sci. 2002, 115, 4661–4669. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lee, J.-A.; Black, D.L. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007, 8, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, W.J.; Chen, C.Y.; Si, Y.; Zhang, X.; Lu, L.; Suswam, E.; Zheng, L.; King, P.H. KSRP: A checkpoint for inflammatory cytokine production in astrocytes. Glia 2012, 60, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Pan, F.; Zhang, H.; Bassell, G.J.; Singer, R.H. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J. Cell Biol. 2002, 156, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Rehbein, M.; Wege, K.; Buck, F.; Schweizer, M.; Richter, D.; Kindler, S. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J. Neurochem. 2002, 82, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Tadessse, H.; Deschenes-Furry, J.; Boisvenue, S.; Côté, J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum. Mol. Genet. 2008, 17, 506–524. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y.; Schneider, R.J. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J. Biol. Chem. 2004, 279, 12974–12979. [Google Scholar] [CrossRef] [PubMed]
- Bird, C.W.; Gardiner, A.S.; Bolognani, F.; Tanner, D.C.; Chen, C.Y.; Lin, W.J.; Yoo, S.; Twiss, J.L.; Perrone-Bizzozero, N. KSRP modulation of GAP-43 mRNA stability restricts axonal outgrowth in embryonic hippocampal neurons. PLoS ONE 2013, 8, e79255. [Google Scholar] [CrossRef] [PubMed]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009, 459, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wan, G.; Berger, F.G.; He, X.; Lu, X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol. Cell 2011, 41, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Michlewski, G.; Caceres, J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010, 17, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.F.; Oleynikov, Y.; Kislauskis, E.H.; Taneja, K.L.; Singer, R.H. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell Biol. 1997, 17, 2158–2165. [Google Scholar] [CrossRef] [PubMed]
- Jonson, L.; Vikesaa, J.; Krogh, A.; Nielsen, L.K.; Hansen, T.; Borup, R.; Johnsen, A.H.; Christiansen, J.; Nielsen, F.C. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell Proteom. 2007, 6, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.L.; Mitra, S.; Harris, R.; Buxbaum, A.R.; Lionnet, T.; Brenowitz, M.; Girvin, M.; Levy, M.; Almo, S.C.; Singer, R.H.; et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012, 26, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Huttelmaier, S.; Zenklusen, D.; Lederer, M.; Dictenberg, J.; Lorenz, M.; Meng, X.; Bassell, G.J.; Condeelis, J.; Singer, R.H. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 2005, 438, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Stohr, N.; Lederer, M.; Reinke, C.; Meyer, S.; Hatzfeld, M.; Singer, R.H.; Huttelmaier, S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006, 175, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, C.J.; Willis, D.E.; Xu, M.; Tep, C.; Jiang, C.; Yoo, S.; Schanen, N.C.; Kirn-Safran, C.B.; van Minnen, J.; English, A.; et al. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011, 30, 4665–4677. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, C.J.; Park, M.; Spillane, M.; Yoo, S.; Pacheco, A.; Gomes, C.; Vuppalanchi, D.; McDonald, M.; Kim, H.H.; Merianda, T.T.; et al. Axonally synthesized beta-actin and GAP-43 proteins support distinct modes of axonal growth. J. Neurosci. 2013, 33, 3311–3322. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Lee, S.J.; Gardiner, A.S.; Perrone-Bizzozero, N.I.; Yoo, S. Different motif requirements for the localization zipcode element of beta-actin mRNA binding by HuD and ZBP1. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villen, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Wang, N.; Houel, S.; Couts, K.; Old, W.; Ahn, N. Dosage and temporal thresholds in microRNA proteomics. Mol. Cell Proteom. 2015, 14, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.; Wen, J.; Marks, D.S.; Krogh, A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010, 20, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Corcoran, D.L.; Nusbaum, J.D.; Reid, D.W.; Georgiev, S.; Hafner, M.; Ascano, M.; Tuschl, T.; Ohler, U.; Keene, J.D. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 2011, 43, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.N.; Habermacher, R.; Martine, U.; Closs, E.I.; Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006, 125, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Srikantan, S.; Abdelmohsen, K.; Lee, E.K.; Tominaga, K.; Subaran, S.S.; Kuwano, Y.; Kulshrestha, R.; Panchakshari, R.; Kim, H.H.; Yang, X.; et al. Translational control of TOP2A influences doxorubicin efficacy. Mol. Cell Biol. 2011, 31, 3790–3801. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, K.; Srikantan, S.; Lee, E.K.; Subaran, S.S.; Martindale, J.L.; Abdelmohsen, K.; Gorospe, M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol. Cell Biol. 2011, 31, 4219–4231. [Google Scholar] [CrossRef] [PubMed]
- Epis, M.R.; Barker, A.; Giles, K.M.; Beveridge, D.J.; Leedman, P.J. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J. Biol. Chem. 2011, 286, 41442–41454. [Google Scholar] [CrossRef] [PubMed]
- Young, L.E.; Moore, A.E.; Sokol, L.; Meisner-Kober, N.; Dixon, D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012, 10, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Connick, M.C.; Vanderhoof, J.; Ishak, M.; Hartley, R.S. MicroRNA-16 modulates HuR regulation of cyclin E1 in breast cancer cells. Int. J. Mol. Sci. 2015, 16, 7112–7132. [Google Scholar] [CrossRef] [PubMed]
- Jing, Q.; Huang, S.; Guth, S.; Zarubin, T.; Motoyama, A.; Chen, J.; di Padova, F.; Lin, S.C.; Gram, H.; Han, J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005, 120, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Glorian, V.; Maillot, G.; Poles, S.; Iacovoni, J.S.; Favre, G.; Vagner, S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011, 18, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Coller, H. Functional interactions between microRNAs and RNA binding proteins. MicroRNA 2012, 1, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Doyle, F.; Tenenbaum, S.A. Trans-regulation of RNA-binding protein motifs by microRNA. Front. Genet. 2014. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, X.; Lei, Y.; Liu, Z.; Tong, T.; Wang, W. Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J. Cell Biochem. 2010, 111, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Hutchison, E.R.; Lee, E.K.; Kuwano, Y.; Kim, M.M.; Masuda, K.; Srikantan, S.; Subaran, S.S.; Marasa, B.S.; Mattson, M.P.; et al. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol. Cell Biol. 2010, 30, 4197–4210. [Google Scholar] [CrossRef] [PubMed]
- Amirouche, A.; Tadesse, H.; Miura, P.; Belanger, G.; Lunde, J.A.; Cote, J.; Jasmin, B.J. Converging pathways involving microRNA-206 and the RNA-binding protein KSRP control post-transcriptionally utrophin A expression in skeletal muscle. Nucleic Acids Res. 2014, 42, 3982–3997. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Gross, C.; Xing, L.; Goshima, Y.; Bassell, G.J. Identification of axon-enriched microRNAs localized to growth cones of cortical neurons. Dev. Neurobiol. 2014, 74, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Natera-Naranjo, O.; Aschrafi, A.; Gioio, A.E.; Kaplan, B.B. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 2010, 18, 1516–1529. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G. MicroRNAs at the synapse. Nat. Rev. Neurosci. 2009, 10, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Perrone-Bizzozero, N.I.; Bolognani, F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J. Neurosci. Res. 2002, 68, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Perrone-Bizzozero, N.I.; Tanner, D.C.; Mounce, J.; Bolognani, F. Increased expression of axogenesis-related genes and mossy fiber length in dentate granule cells from adult HuD overexpressor mice. ASN Neuro. 2011, 3, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Merianda, T.T.; Lee, S.J.; Yoo, S.; Twiss, J.L. Molecular determinants of the axonal mRNA transcriptome. Dev. Neurobiol. 2014, 74, 218–232. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gardiner, A.S.; Twiss, J.L.; Perrone-Bizzozero, N.I. Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function. Biomolecules 2015, 5, 2903-2918. https://doi.org/10.3390/biom5042903
Gardiner AS, Twiss JL, Perrone-Bizzozero NI. Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function. Biomolecules. 2015; 5(4):2903-2918. https://doi.org/10.3390/biom5042903
Chicago/Turabian StyleGardiner, Amy S., Jeffery L. Twiss, and Nora I. Perrone-Bizzozero. 2015. "Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function" Biomolecules 5, no. 4: 2903-2918. https://doi.org/10.3390/biom5042903