A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets
Abstract
1. Introduction
1.1. Structure of SIRT6 and its Activity
1.2. Observed Implications of SIRT6
2. Role of SIRT6 in DNA Repair and Aging-Related Processes
2.1. Maintenance of Telomeres
2.2. Base Excision Repair
2.3. Double Strand DNA Break Repair
2.4. Aging and Life Expectancy
3. SIRT6 in Glucose Metabolism and Diabetes
3.1. SIRT6 in Glycolysis and the Suppression of Glucose-Metabolic Genes
3.2. SIRT6 in Gluconeogenesis
4. Pharmacological Intervention with Small Molecule SIRT6 Inhibitors for SIRT Associated Ailments
5. SIRT6 in Heart Disease
6. SIRT6 and Cancer
6.1. Tumor Growth Resulting from Altered Glycolysis
6.2. SIRT6 and the Initiation of Hepatic Cancer
6.3. Aberrant Behavior of SIRT6 in Other Forms of Cancer
7. SIRT6 in Neurodegenerative Diseases and Brain Aging
8. Advances Made with SIRT6 in Other Areas
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Frye, R.A. Phylogenetic Classification of Prokaryotic and Eukaryotic Sir2-like Proteins. Biochem. Biophys. Res. Commun. 2000, 273, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Klar, A.J.S.; Strathern, J.N.; Broach, J.R.; Hicks, J.B. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature 1981, 289, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999, 13, 2570–2580. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, R.; Faulkner, S.; Miyokawa, R.; Burhenn, L.; Henriksen, M.; Wood, J.G.; Helfand, S.L. Increased expression of Drosophila Sir 2 extends life span in a dose-dependent manner. Aging (Albany. N. Y.) 2013, 5, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Moynihan, K.A.; Grimm, A.A.; Plueger, M.M.; Bernal-Mizrachi, E.; Ford, E.; Cras-Méneur, C.; Permutt, M.A.; Imai, S. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005, 2, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Bryk, M.; Banerjee, M.; Murphy, M.; Knudsen, K.E.; Garfinkel, D.J.; Curcio, M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997, 11, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Boeke, J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997, 11, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, S.; Esposito, R.E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 1989, 56, 771–776. [Google Scholar] [CrossRef]
- Rogina, B.; Helfand, S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 2004, 101, 15998–16003. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T.; Deng, C.-X.; Mostoslavsky, R. Recent progress in the biology and physiology of sirtuins. Nature 2009, 460, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Hallows, W.C.; Yu, W.; Smith, B.C.; Devires, M.K.; Ellinger, J.J.; Someya, S.; Shortreed, M.R.; Prolla, T.; Markley, J.L.; Smith, L.M.; et al. Sirt3 Promotes the Urea Cycle and Fatty Acid Oxidation during Dietary Restriction. Mol. Cell 2011, 41, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Someya, S.; Yu, W.; Hallows, W.C.; Xu, J.; Vann, J.M.; Leeuwenburgh, C.; Tanokura, M.; Denu, J.M.; Prolla, T.A. Sirt3 Mediates Reduction of Oxidative Damage and Prevention of Age-Related Hearing Loss under Caloric Restriction. Cell 2010, 143, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Lomb, D.J.; Haigis, M.C.; Guarente, L. SIRT5 Deacetylates Carbamoyl Phosphate Synthetase 1 and Regulates the Urea Cycle. Cell 2009, 137, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 2005, 16, 4623–4635. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Brown, K.; Hirschey, M.D.; Verdin, E.; Chen, D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metab. 2010, 12, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Coleman, M.C.; Pennington, J.D.; Ozden, O.; Park, S.-H.; Jiang, H.; Kim, H.-S.; Flynn, C.R.; Hill, S.; Hayes McDonald, W.; et al. Sirt3-Mediated Deacetylation of Evolutionarily Conserved Lysine 122 Regulates MnSOD Activity in Response to Stress. Mol. Cell 2010, 40, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Hebert, A.S.; Dittenhafer-Reed, K.E.; Yu, W.; Bailey, D.J.; Selen, E.S.; Boersma, M.D.; Carson, J.J.; Tonelli, M.; Balloon, A.J.; Higbee, A.J.; et al. Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome. Mol. Cell 2013, 49, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Mostoslavsky, R.; Chua, K.F.; Lombard, D.B.; Pang, W.W.; Fischer, M.R.; Gellon, L.; Liu, P.; Mostoslavsky, G.; Franco, S.; Murphy, M.M.; et al. Genomic Instability and Aging-like Phenotype in the Absence of Mammalian SIRT6. Cell 2006, 124, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, K.; Zhang, N.; Amador, G.; Wang, Y.; Zhao, S.; Li, L.; Qiu, Y.; Wang, Z. Overexpressed SIRT6 attenuates cisplatin-induced acute kidney injury by inhibiting ERK1/2 signaling. Kidney Int. 2018, 93, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Kugel, S.; Mostoslavsky, R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 2014, 39, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Wang, Z.; Ma, C.; Jiang, Y.; Zhang, N.; Hu, K.; Li, L.; Wang, Z. Metformin promotes the proliferation and differentiation of murine preosteoblast by regulating the expression of sirt6 and oct4. Pharmacol. Res. 2018, 129, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Kanfi, Y.; Shalman, R.; Peshti, V.; Pilosof, S.N.; Gozlan, Y.M.; Pearson, K.J.; Lerrer, B.; Moazed, D.; Marine, J.-C.; de Cabo, R.; et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008, 582, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Elhanati, S.; Kanfi, Y.; Varvak, A.; Roichman, A.; Carmel-Gross, I.; Barth, S.; Gibor, G.; Cohen, H.Y. Multiple Regulatory Layers of SREBP1/2 by SIRT6. Cell Rep. 2013, 4, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.-X.; Dong, X.C. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 2013, 54, 2745–2753. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.M.; Xiao, C.; Finley, L.W.S.; Lahusen, T.; Souza, A.L.; Pierce, K.; Li, Y.-H.; Wang, X.; Laurent, G.; German, N.J.; et al. SIRT4 Has Tumor-Suppressive Activity and Regulates the Cellular Metabolic Response to DNA Damage by Inhibiting Mitochondrial Glutamine Metabolism. Cancer Cell 2013, 23, 450–463. [Google Scholar] [CrossRef] [PubMed]
- Onyango, P.; Celic, I.; McCaffery, J.M.; Boeke, J.D.; Feinberg, A.P. SIRT3, a human SIR2 homologue, is an NAD- dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. USA 2002, 99, 13653–13658. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Liu, B.; Wang, Y.; Hao, Q.; Zhou, Z. Lamin A Is an Endogenous SIRT6 Activator and Promotes SIRT6-Mediated DNA Repair. Cell Rep. 2015, 13, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, M.; Zullo, A.; Servillo, L.; Mancini, F.P.; Borriello, A.; Giovane, A.; Della Ragione, F.; D’Onofrio, N.; Balestrieri, M.L. Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res. Rev. 2017, 35, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.; Rymarchyk, S.; Zheng, S.; Cen, Y. Trichostatin A inhibits deacetylation of histone H3 and p53 by SIRT6. Arch. Biochem. Biophys. 2018, 638, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zwaans, B.M.M.; Eckersdorff, M.; Lombard, D.B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 2009, 8, 2662–2663. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Dittenhafer-Reed, K.E.; Denu, J.M. Sirtuin Catalysis and Regulation. J. Biol. Chem. 2012, 287, 42419–42427. [Google Scholar] [CrossRef] [PubMed]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.D.; Denu, J.M. Structural Identification of 2′- and 3′-O-Acetyl-ADP-ribose as Novel Metabolites Derived from the Sir2 Family of β-NAD+-dependent Histone/Protein Deacetylases. J. Biol. Chem. 2002, 277, 18535–18544. [Google Scholar] [CrossRef] [PubMed]
- Sauve, A.A.; Celic, I.; Avalos, J.; Deng, H.; Boeke, J.D.; Schramm, V.L. Chemistry of gene silencing: The mechanism of NAD+-dependent deacetylation reactions. Biochemistry 2001, 40, 15456–15463. [Google Scholar] [CrossRef] [PubMed]
- Haigis, M.C.; Guarente, L.P. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006, 20, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.W.; Feldman, J.L.; Devries, M.K.; Dong, A.; Edwards, A.M.; Denu, J.M. Structure and Biochemical Functions of SIRT6. J. Biol. Chem. 2011, 286, 14575–14587. [Google Scholar] [CrossRef] [PubMed]
- Damonte, P.; Sociali, G.; Parenti, M.D.; Soncini, D.; Bauer, I.; Boero, S.; Grozio, A.; Holtey, M.V.; Piacente, F.; Becherini, P.; Sanguineti, R.; et al. SIRT6 inhibitors with salicylate-like structure show immunosuppressive and chemosensitizing effects. Bioorg. Med. Chem. 2017, 25, 5849–5858. [Google Scholar] [CrossRef] [PubMed]
- Finnin, M.S.; Donigian, J.R.; Pavletich, N.P. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 2001, 8, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wei, W.; Jiang, Y.; Peng, H.; Cai, J.; Mao, C.; Dai, H.; Choy, W.; Bemis, J.E.; Jirousek, M.R.; et al. Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes. J. Biol. Chem. 2009, 284, 24394–24405. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.; Min, J.; Antoshenko, T.; Wang, C.-L.; Allali-Hassani, A.; Dong, A.; Loppnau, P.; Vedadi, M.; Bochkarev, A.; Sternglanz, R.; et al. Structural Basis of Inhibition of the Human NAD+-Dependent Deacetylase SIRT5 by Suramin. Structure 2007, 15, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, A.; Scher, M.; Lee, D.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Human SirT1 Interacts with Histone H1 and Promotes Formation of Facultative Heterochromatin. Mol. Cell 2004, 16, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Anamika, K.; Abhinandan, K.R.; Deshmukh, K.; Srinivasan, N. Classification of nonenzymatic homologues of protein kinases. Comp. Funct. Genom. 2009, 2009, 365637. [Google Scholar] [CrossRef] [PubMed]
- Pils, B.; Schultz, J. Evolution of the Multifunctional Protein Tyrosine Phosphatase Family. Mol. Biol. Evol. 2004, 21, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Tanner, K.G.; Landry, J.; Sternglanz, R.; Denu, J.M. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 2000, 97, 14178–14182. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Hine, C.; Tian, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 Promotes DNA Repair Under Stress by Activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef] [PubMed]
- Tennen, R.I.; Berber, E.; Chua, K.F. Functional dissection of SIRT6: Identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 2010, 131, 185–192. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Criscione, S.W.; Peckham, E.J.; Hillenmeyer, S.; Hamm, E.A.; Manivannan, J.; Peterson, A.L.; Kreiling, J.A.; Neretti, N.; Sedivy, J.M. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 2013, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Criscione, S.W.; Peterson, A.L.; Neretti, N.; Sedivy, J.M.; Kreiling, J.A. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany. N. Y.) 2013, 5, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Diecke, S.; Zhang, W.Y.; Lan, F.; He, C.; Mordwinkin, N.M.; Chua, K.F.; Wu, J.C. The Role of SIRT6 Protein in Aging and Reprogramming of Human Induced Pluripotent Stem Cells. J. Biol. Chem. 2013, 288, 18439–18447. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.E.; Gomes, A.P.; Sinclair, D.A. Geroncogenesis: Metabolic Changes during Aging as a Driver of Tumorigenesis. Cancer Cell 2014, 25, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Mao, Z.; Hine, C. Changes in DNA repair during aging. Nucleic Acids Res. 2007, 35, 7466–7474. [Google Scholar] [CrossRef] [PubMed]
- Orkin, S.H.; Hochedlinger, K. Chromatin connections to pluripotency and cellular reprogramming. Cell 2011, 145, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Tennen, R.I.; Chua, K.F. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem. Sci. 2011, 36, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, N.; Zhang, H.; Zhang, H.; Qiao, J.; Jia, W.; Zhu, S.; Mao, Z.; Kang, J. Sirt6 Promotes DNA End Joining in iPSCs Derived from Old Mice. Cell Rep. 2017, 18, 2880–2892. [Google Scholar] [CrossRef] [PubMed]
- McCord, R.A.; Michishita, E.; Hong, T.; Berber, E.; Boxer, L.D.; Kusumoto, R.; Guan, S.; Shi, X.; Gozani, O.; Burlingame, A.L.; et al. SIRT6 stabilizes DNA-dependent Protein Kinase at chromatin for DNA double-strand break repair. Aging (Albany. N. Y.) 2009, 1, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.A.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; McCord, R.A.; Boxer, L.D.; Barber, M.F.; Hong, T.; Gozani, O.; Chua, K.F. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009, 8, 2664–2666. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, S.; Lee, L.; Davalos, A.; Schiestl, R.H.; Campisi, J.; Oshima, J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell 2003, 2, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Multani, A.S.; Chang, S. WRN at telomeres: implications for aging and cancer. J. Cell Sci. 2007, 120, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; de Lange, T. Mammalian telomeres end in a large duplex loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Murti, K.G.; Prescott, D.M. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl. Acad. Sci. USA 1999, 96, 14436–14439. [Google Scholar] [CrossRef] [PubMed]
- Smogorzewska, A.; de Lange, T. Regulation of Telomerase by Telomeric Proteins. Annu. Rev. Biochem. 2004, 73, 177–208. [Google Scholar] [CrossRef] [PubMed]
- Kaidi, A.; Weinert, B.T.; Choudhary, C.; Jackson, S.P. Human SIRT6 Promotes DNA End Resection Through CtIP Deacetylation. Science 2010, 329, 1348–1353. [Google Scholar] [CrossRef] [PubMed]
- Andres, S.N.; Williams, R.S. CtIP/Ctp1/Sae2, molecular form fit for function. DNA Repair (Amst). 2017, 56, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Toiber, D.; Erdel, F.; Bouazoune, K.; Silberman, D.M.; Zhong, L.; Mulligan, P.; Sebastian, C.; Cosentino, C.; Martinez-Pastor, B.; Giacosa, S.; et al. SIRT6 Recruits SNF2H to DNA Break Sites, Preventing Genomic Instability through Chromatin Remodeling. Mol. Cell 2013, 51, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Tasselli, L.; Zheng, W.; Chua, K.F. SIRT6: Novel Mechanisms and Links to Aging and Disease. Trends Endocrinol. Metab. 2017, 28, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Satoh, A.; Brace, C.S.; Rensing, N.; Cliften, P.; Wozniak, D.F.; Herzog, E.D.; Yamada, K.A.; Imai, S. Sirt1 Extends Life Span and Delays Aging in Mice through the Regulation of Nk2 Homeobox 1 in the DMH and LH. Cell Metab. 2013, 18, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Rizki, G.; Iwata, T.N.; Li, J.; Riedel, C.G.; Picard, C.L.; Jan, M.; Murphy, C.T.; Lee, S.S. The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO. PLoS Genet. 2011, 7, e1002235. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, M.; Guarente, L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature 2011, 477, E1–E2. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Tsaih, S.-W.; Petkova, S.B.; De Evsikova, C.M.; Xing, S.; Marion, M.A.; Bogue, M.A.; Mills, K.D.; Peters, L.L.; Bult, C.J.; et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell 2009, 8, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Estep, P.W.; Warner, J.B.; Bulyk, M.L. Short-Term Calorie Restriction in Male Mice Feminizes Gene Expression and Alters Key Regulators of Conserved Aging Regulatory Pathways. PLoS ONE 2009, 4, e5242. [Google Scholar] [CrossRef] [PubMed]
- Selman, C.; Kerrison, N.D.; Cooray, A.; Piper, M.D.W.; Lingard, S.J.; Barton, R.H.; Schuster, E.F.; Blanc, E.; Gems, D.; Nicholson, J.K.; et al. Coordinated multitissue transcriptional and plasma metabonomic profiles following acute caloric restriction in mice. Physiol. Genom. 2006, 27, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. Stress granules. Curr. Biol. 2009, 19, R397–R398. [Google Scholar] [CrossRef] [PubMed]
- Jedrusik-Bode, M.; Studencka, M.; Smolka, C.; Baumann, T.; Schmidt, H.; Kampf, J.; Paap, F.; Martin, S.; Tazi, J.; Muller, K.M.; et al. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J. Cell Sci. 2013, 126, 5166–5177. [Google Scholar] [CrossRef] [PubMed]
- Bluher, M.; Kahn, B.B.; Kahn, C.R. Extended Longevity in Mice Lacking the Insulin Receptor in Adipose Tissue. Science 2003, 299, 572–574. [Google Scholar] [CrossRef] [PubMed]
- Conti, B.; Sanchez-Alavez, M.; Winsky-Sommerer, R.; Morale, M.C.; Lucero, J.; Brownell, S.; Fabre, V.; Huitron-Resendiz, S.; Henriksen, S.; Zorrilla, E.P.; et al. Transgenic Mice with a Reduced Core Body Temperature Have an Increased Life Span. Science 2006, 314, 825–828. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Herranz, D.; Muñoz-Martin, M.; Cañamero, M.; Mulero, F.; Martinez-Pastor, B.; Fernandez-Capetillo, O.; Serrano, M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat. Commun. 2010, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Wang, G.; Tao, R.; Wu, P.; Kono, T.; Li, K.; Ding, W.-X.; Tong, X.; Tersey, S.A.; Harris, R.A.; et al. Sirtuin 6 regulates glucose-stimulated insulin secretion in mouse pancreatic beta cells. Diabetologia 2016, 59, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Sebastián, C.; Zwaans, B.M.M.; Silberman, D.M.; Gymrek, M.; Goren, A.; Zhong, L.; Ram, O.; Truelove, J.; Guimaraes, A.R.; Toiber, D.; et al. The Histone Deacetylase SIRT6 Is a Tumor Suppressor that Controls Cancer Metabolism. Cell 2012, 151, 1185–1199. [Google Scholar] [CrossRef] [PubMed]
- Dominy, J.E.; Lee, Y.; Jedrychowski, M.P.; Chim, H.; Jurczak, M.J.; Camporez, J.P.; Ruan, H.-B.; Feldman, J.; Pierce, K.; Mostoslavsky, R.; et al. The Deacetylase Sirt6 Activates the Acetyltransferase GCN5 and Suppresses Hepatic Gluconeogenesis. Mol. Cell 2012, 48, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P.; Rhee, J.; Donovan, J.; Walkey, C.J.; Yoon, J.C.; Oriente, F.; Kitamura, Y.; Altomonte, J.; Dong, H.; Accili, D.; et al. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature 2003, 423, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Tao, R.; DePinho, R.A.; Dong, X.C. Deletion of Hepatic FoxO1/3/4 Genes in Mice Significantly Impacts on Glucose Metabolism through Downregulation of Gluconeogenesis and Upregulation of Glycolysis. PLoS ONE 2013, 8, e74340. [Google Scholar] [CrossRef] [PubMed]
- Parenti, M.D.; Grozio, A.; Bauer, I.; Galeno, L.; Damonte, P.; Millo, E.; Sociali, G.; Franceschi, C.; Ballestrero, A.; Bruzzone, S.; Del Rio, A.; Nencioni, A. Discovery of Novel and Selective SIRT6 Inhibitors. J. Med. Chem. 2014, 57, 4796–4804. [Google Scholar] [CrossRef] [PubMed]
- Sociali, G.; Magnone, M.; Ravera, S.; Damonte, P.; Vigliarolo, T.; Von Holtey, M.; Vellone, V.G.; Millo, E.; Caffa, I.; Cea, M.; et al. Pharmacological Sirt6 inhibition improves glucose tolerance in a type 2 diabetes mouse model. FASEB J. 2017, 31, 3138–3149. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Choi, J.M.; Chang, E.; Park, S.W.; Park, C.-Y. Sirt1 and Sirt6 Mediate Beneficial Effects of Rosiglitazone on Hepatic Lipid Accumulation. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Chalkiadaki, A.; Guarente, L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 2012, 8, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Song, H.Y.; Rellinger, E.J.; Park, S.-H.; Paul, P.; Qiao, J.; Vasilopoulos, A.; Ozden, O.; Gius, D.; Chung, D.H. Inhibition of Sirtuin 6 Induces Neuroblastoma Differentiation. Anticancer Res. 2018, 38, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, S.-S.; Chen, S.-R.; Pi, R.-B.; Gao, S.; Li, H.; Ye, J.-T.; Liu, P.-Q. Nmnat2 protects cardiomyocytes from hypertrophy via activation of SIRT6. FEBS Lett. 2012, 586, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, N.R.; Vasudevan, P.; Zhong, L.; Kim, G.; Samant, S.; Parekh, V.; Pillai, V.B.; Ravindra, P.V.; Gupta, M.; Jeevanandam, V.; et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 2012, 18, 1643–1650. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, D.; Liu, Z.; Li, M.; Hong, H.; Liu, C.; Gao, S.; Li, H.; Cai, Y.; Chen, S.; et al. SIRT6 suppresses isoproterenol-induced cardiac hypertrophy through activation of autophagy. Transl. Res. 2016, 172, 96–112. [Google Scholar] [CrossRef] [PubMed]
- Metallo, C.M.; Vander Heiden, M.G. Understanding metabolic regulation and its influence on cell physiology. Mol. Cell 2013, 49, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. THE METABOLISM OF TUMORS IN THE BODY. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Karnezis, A.N.; Garcia, D.; Madriles, F.; Kortlever, R.M.; Rostker, F.; Brown Swigart, L.; Pham, D.M.; Seo, Y.; Evan, G.I.; et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 2010, 468, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Pikarsky, E.; Porat, R.M.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Montorsi, M.; Maggioni, M.; Falleni, M.; Pellegrini, C.; Donadon, M.; Torzilli, G.; Santambrogio, R.; Spinelli, A.; Coggi, G.; Bosari, S. Survivin gene expression in chronic liver disease and hepatocellular carcinoma. Hepatogastroenterology 2007, 54, 2040–2044. [Google Scholar]
- Park, E.J.; Lee, J.H.; Yu, G.-Y.; He, G.; Ali, S.R.; Holzer, R.G.; Österreicher, C.H.; Takahashi, H.; Karin, M. Dietary and Genetic Obesity Promote Liver Inflammation and Tumorigenesis by Enhancing IL-6 and TNF Expression. Cell 2010, 140, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Meixner, A.; Karreth, F.; Kenner, L.; Penninger, J.M.; Wagner, E.F. Jun and JunD-dependent functions in cell proliferation and stress response. Cell Death Differ. 2010, 17, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Ji, Y.; Bakiri, L.; Qiu, Z.; Cen, J.; Chen, X.; Chen, L.; Scheuch, H.; Zheng, H.; Qin, L.; et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat. Cell Biol. 2012, 14, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-C.; Lin, P.-M.; Lin, S.-F.; Hsu, C.-H.; Lin, H.-C.; Hu, M.-L.; Hsu, C.-M.; Yang, M.-Y. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumor Biol. 2013, 34, 1847–1854. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Kafeel, M.I.; Avezbakiyev, B.; Chen, C.; Sun, Y.; Rathnasabapathy, C.; Kalavar, M.; He, Z.; Burton, J.; Lichter, S. Histone Deacetylase in Chronic Lymphocytic Leukemia. Oncology 2011, 81, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Cagnetta, A.; Soncini, D.; Orecchioni, S.; Talarico, G.; Minetto, P.; Guolo, F.; Retali, V.; Colombo, N.; Carminati, E.; Clavio, M.; et al. Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells’ vulnerability to DNA-damaging agents. Haematologica 2018, 103, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Cea, M.; Cagnetta, A.; Adamia, S.; Acharya, C.; Tai, Y.-T.; Fulciniti, M.; Ohguchi, H.; Munshi, A.; Acharya, P.; Bhasin, M.K.; et al. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood 2016, 127, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Jęśko, H.; Wencel, P.; Strosznajder, R.P.; Strosznajder, J.B. Sirtuins and Their Roles in Brain Aging and Neurodegenerative Disorders. Neurochem. Res. 2017, 42, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Braidy, N.; Poljak, A.; Grant, R.; Jayasena, T.; Mansour, H.; Chan-Ling, T.; Smythe, G.; Sachdev, P.; Guillemin, G.J. Differential expression of sirtuins in the aging rat brain. Front. Cell. Neurosci. 2015, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.S.; Choi, H.; Song, H.; Hwang, Y.J.; Kim, A.; Ryu, H.; Mook-Jung, I. p53-dependent SIRT6 expression protects Aβ42-induced DNA damage. Sci. Rep. 2016, 6, 25628. [Google Scholar] [CrossRef] [PubMed]
- Kaluski, S.; Portillo, M.; Besnard, A.; Stein, D.; Einav, M.; Zhong, L.; Ueberham, U.; Arendt, T.; Mostoslavsky, R.; Sahay, A.; et al. Neuroprotective Functions for the Histone Deacetylase SIRT6. Cell Rep. 2017, 18, 3052–3062. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wei, R.; Zhang, L.; Tan, Y.; Qian, C. Sirtuin 6 protects the brain from cerebral ischemia/reperfusion injury through NRF2 activation. Neuroscience 2017, 366, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Xiao, C.; Wang, R.-H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.-I.; Park, O.; Ki, S.H.; et al. Hepatic-Specific Disruption of SIRT6 in Mice Results in Fatty Liver Formation Due to Enhanced Glycolysis and Triglyceride Synthesis. Cell Metab. 2010, 12, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.-X.; Dong, X.C. FoxO3 Transcription Factor and Sirt6 Deacetylase Regulate Low Density Lipoprotein (LDL)-cholesterol Homeostasis via Control of the Proprotein Convertase Subtilisin/Kexin Type 9 (Pcsk9) Gene Expression. J. Biol. Chem. 2013, 288, 29252–29259. [Google Scholar] [CrossRef] [PubMed]
- Van Gool, F.; Gallí, M.; Gueydan, C.; Kruys, V.; Prevot, P.-P.; Bedalov, A.; Mostoslavsky, R.; Alt, F.W.; De Smedt, T.; Leo, O. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat. Med. 2009, 15, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.L.A.; Michishita, E.; Adler, A.S.; Damian, M.; Berber, E.; Lin, M.; McCord, R.A.; Ongaigui, K.C.L.; Boxer, L.D.; Chang, H.Y.; et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-κB-Dependent Gene Expression and Organismal Life Span. Cell 2009, 136, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Bauer, I.; Grozio, A.; Lasigliè, D.; Basile, G.; Sturla, L.; Magnone, M.; Sociali, G.; Soncini, D.; Caffa, I.; Poggi, A.; et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J. Biol. Chem. 2012, 287, 40924–40937. [Google Scholar] [CrossRef] [PubMed]
- Magnone, M.; Bauer, I.; Poggi, A.; Mannino, E.; Sturla, L.; Brini, M.; Zocchi, E.; De Flora, A.; Nencioni, A.; Bruzzone, S. NAD+ Levels Control Ca2+ Store Replenishment and Mitogen-induced Increase of Cytosolic Ca2+ by Cyclic ADP-ribose-dependent TRPM2 Channel Gating in Human T Lymphocytes. J. Biol. Chem. 2012, 287, 21067–21081. [Google Scholar] [CrossRef] [PubMed]
- Sugatani, T.; Agapova, O.; Malluche, H.H.; Hruska, K.A. SIRT6 deficiency culminates in low-turnover osteopenia. Bone 2015, 81, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Rahnasto-Rilla, M.; Tyni, J.; Huovinen, M.; Jarho, E.; Kulikowicz, T.; Ravichandran, S.; Vilhelm, A.B.; Ferrucci, L.; Lahtela-Kakkonen, M.; Moaddel, R. Natural polyphenols as sirtuin 6 modulators. Sci. Rep. 2018, 8, 4163. [Google Scholar] [CrossRef] [PubMed]
- Kokkonen, P.; Rahnasto-Rilla, M.; Mellini, P.; Jarho, E.; Lahtela-Kakkonen, M.; Kokkola, T. Studying SIRT6 regulation using H3K56 based substrate and small molecules. Eur. J. Pharm. Sci. 2014, 63, 71–76. [Google Scholar] [CrossRef] [PubMed]



| Substrate | Linked Functions in Cell |
|---|---|
| H3K9ac | Regulation of transcription, stability of telomeres, response to DNA damage |
| H3K56ac | Regulation of transcription, stability of telomeres, response to DNA damage |
| H3K18ac | Silencing of heterochromatin |
| NPM1 | Cellular Senescence |
| PARP1 | DNA double-strand break repair and base-excision repair |
| KAP1 | DNA double-strand break repair |
| Asinex ID | Compound Structure | Percentage Inhibition of SIRT6 at 200 µM Concentration of the Compound |
|---|---|---|
| SYN17739303 | 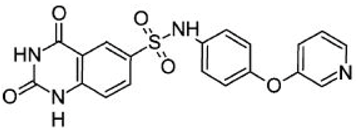 | 100 ± 4 |
| BAS13555470 | 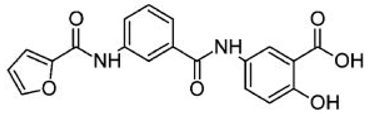 | 62 ± 7 |
| SYN10366754 | 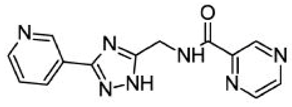 | 12 ± 3 |
| BAS00417531 | 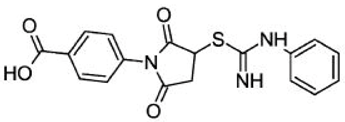 | 66 ± 6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, R.I.; Nirzhor, S.S.R.; Akter, R. A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules 2018, 8, 44. https://doi.org/10.3390/biom8030044
Khan RI, Nirzhor SSR, Akter R. A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules. 2018; 8(3):44. https://doi.org/10.3390/biom8030044
Chicago/Turabian StyleKhan, Rubayat Islam, Saif Shahriar Rahman Nirzhor, and Raushanara Akter. 2018. "A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets" Biomolecules 8, no. 3: 44. https://doi.org/10.3390/biom8030044
APA StyleKhan, R. I., Nirzhor, S. S. R., & Akter, R. (2018). A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules, 8(3), 44. https://doi.org/10.3390/biom8030044




