Multi-Spectroscopic and Theoretical Analysis on the Interaction between Human Serum Albumin and a Capsaicin Derivative—RPF101
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Steady-State Fluorescence Measurements
2.3. Time-Resolved Fluorescence Measurements
2.4. Synchronous Fluorescence Measurements
2.5. Zeta Potential Measurements
2.6. Circular Dichroism Measurements
2.7. Drug Displacement Experiment
2.8. Molecular Docking Studies
3. Results
3.1. Steady-State and Time-Resolved Fluorescence Quenching
3.2. Synchronous Fluorescence Spectroscopy
3.3. Zeta Potential Studies
3.4. Change on the Protein Secondary Structure Induced by RPF101 Binding
3.5. Competitive Binding Studies
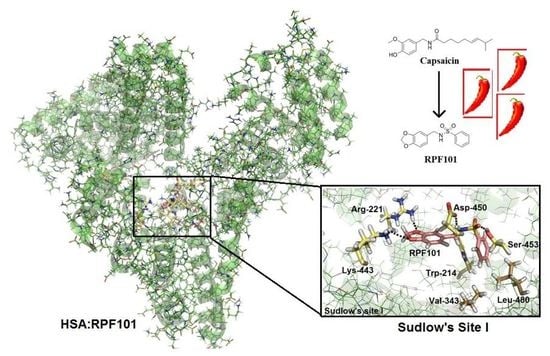
3.6. Molecular Docking Studies for the Interaction HSA:RPF101
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clark, R.; Lee, S. Anticancer properties of capsaicin against human cancer. Anticancer Res. 2016, 36, 837–844. [Google Scholar] [PubMed]
- Lee, J.H.; Lee, Y.; Ryu, H.C.; Kang, D.W.; Lee, J.; Lazar, J.; Pearce, L.V.; Pavlyukovets, V.A.; Blumberg, P.M.; Choi, S. Structural insights into transient receptor potential vanilloid type 1 (TRPV1) from homology modeling, flexible docking, and mutational studies. J. Comput. Aided Mol. Des. 2011, 25, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.L.; Tan, L.T.H.; Ming, L.C.; Chan, K.G.; Lee, L.H.; Goh, B.H.; Khan, T.M. The effectiveness and safety of topical capsaicin in postherpetic neuralgia: A systematic review and meta-analysis. Front. Pharmacol. 2017, 7, 538. [Google Scholar] [CrossRef] [PubMed]
- Sá-Júnior, P.L.; Pasqualoto, K.F.M.; Ferreira, A.K.; Tavares, M.T.; Damião, M.C.F.C.B.; De Azevedo, R.A.; Câmara, D.A.D.; Pereira, A.; Souza, D.M.; Parise-Filho, R. RPF101, a new capsaicin-like analogue, disrupts the microtubule network accompanied by arrest in the G2/M phase, inducing apoptosis and mitotic catastrophe in the MCF-7 breast cancer cells. Toxicol. Appl. Pharmacol. 2013, 266, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Damião, M.C.; Pasqualoto, K.F.; Ferreira, A.K.; Teixeira, S.F.; Azevedo, R.A.; Barbuto, J.A.; Berl, F.P.; Franchi-Junior, G.C.; Nowill, A.E.; Tavares, M.T.; et al. Novel capsaicin analogues as potential anticancer agents: Synthesis, biological evaluation, and In silico approach. Archiv der Pharmazie 2014, 347, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Tavares, M.T.; Pasqualoto, K.F.M.; Van de Streek, J.; Ferreira, A.K.; Azevedo, R.A.; Damião, M.C.F.C.B.; Rodrigues, C.P.; de-Sá-Júnior, P.L.; Barbuto, J.A.M.; Parise-Filho, R.; et al. Synthesis, characterization, in silico approach and in vitro antiproliferative activity of RPF151, a benzodioxole sulfonamide analogue designed from capsaicin scaffold. J. Mol. Struct. 2015, 1088, 138–146. [Google Scholar] [CrossRef]
- Ferreira, A.K.; Tavares, M.T.; Pasqualoto, K.F.M.; Azevedo, R.A.; Teixeira, S.F.; Ferreira-Junior, W.A.; Bertin, A.M.; de-Sá-Junior, P.L.; Barbuto, J.A.M.; Figueiredo, C.R.; et al. RPF151, a novel capsaicin-like analogue: In vitro studies and in vivo preclinical antitumor evaluation in a breast cancer model. Tumor Biol. 2015, 36, 7251–7267. [Google Scholar] [CrossRef] [PubMed]
- Peters, T. Serum albumin. Adv. Protein Chem. 1985, 37, 161–245. [Google Scholar] [PubMed]
- Yeggoni, D.P.; Gokara, M.; Manidhar, D.M.; Rachamallu, A.; Nakka, S.; Reddy, C.S.; Subramanyam, R. Binding and molecular dynamics studies of 7-hydroxycoumarin derivatives with human serum albumin and its pharmacological importance. Mol. Pharm. 2014, 11, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Carter, D.C.; Ho, J.X. Structure of serum albumin. Adv. Protein Chem. 1994, 45, 153–203. [Google Scholar] [PubMed]
- Carter, D.C.; He, X.-M.; Munson, S.H.; Twigg, P.D.; Gernert, K.M.; Broom, M.B.; Miller, T.Y. Three-dimensional structure of human serum albumin. Science 1989, 244, 1195–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahsavani, M.B.; Ahmadi, S.; Aseman, M.D.; Nabavizadeh, S.M.; Alavianmehr, M.M.; Yousef, R. Comparative study on the interaction of two binuclear Pt (II) complexes with human serum albumin: Spectroscopic and docking simulation assessments. J. Photochem. Photobiol. B Biol. 2016, 164, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Kratz, F. Albumin as a drug carrier: Design of prodrugs, drug conjugates and nanoparticles. J. Control. Release 2008, 132, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Danhier, F.; Feron, O.; Préat, V. To exploit the tumor microenvironment: Passive and active tumor targeting nanocarriers for anti-cancer drug delivery. J. Control. Release 2010, 148, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Trieu, V.; Yao, Z.; Louie, L.; Ci, S.; Yang, A.; Tao, C.; De, T.; Beals, B.; Dykes, D.; et al. Increased antitumor activity, intratumor paclitaxel concentrations, and endothelial cell transport of cremophor-free, albumin-bound paclitaxel, ABI-007, compared with cremophor-based paclitaxel. Clin. Cancer Res. 2006, 12, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Ai, N.; Xu, D.; Fan, X. Exploring the interaction between Salvia miltiorrhiza and human serum albumin: Insights from herb—drug interaction reports, computational analysis and experimental studies. Spectrochim. Acta Mol. Biomol. Spectrosc. 2016, 161, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chaves, O.A.; Jesus, C.S.H.; Cruz, P.F.; Sant’Anna, C.M.R.; Brito, R.M.M.; Serpa, C. Evaluation by fluorescence, STD-NMR, docking and semi-empirical calculations of the o-NBA photo-acid interaction with BSA. Spectrochim. Acta Mol. Biomol. Spectrosc. 2016, 169, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Xu, H.; Cao, Y.; Wang, F.; Mi, W. Elucidating the interaction of propofol and serum albumin by spectroscopic and docking methods. J. Mol. Liq. 2016, 219, 405–410. [Google Scholar] [CrossRef]
- Wardell, M.; Wang, Z.; Ho, J.X.; Robert, J.; Ruker, F.; Ruble, J.; Carter, D.C. The atomic structure of human methemalbumin at 1.9 Å. Biochem. Biophys. Res. Commun. 2002, 291, 813–819. [Google Scholar] [CrossRef] [PubMed]
- The Cambridge Crystallographic Data Centre (CCDC). Available online: http://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/ (accessed on 5 July 2018).
- Chaves, O.A.; Cesarin-Sobrinho, D.; Sant’Anna, C.M.R.; de Carvalho, M.G.; Suzart, L.R.; Catunda-Junior, F.E.A.; Netto-Ferreira, J.C.; Ferreira, A.B.B. Probing the interaction between 7-O-β-d-glucopyranosyl-6-(3-methylbut-2-enyl)-5,4’-dihydroxyflavonol with bovine serum albumin (BSA). J. Photochem. Photobiol. A Chem. 2017, 336, 32–41. [Google Scholar] [CrossRef]
- Chaves, O.A.; Santos, M.R.L.; Oliveira, M.C.C.; Sant’Anna, C.M.R.; Ferreira, R.C.; Echevarria, A.; Netto-Ferreira, J.C. Synthesis, tyrosinase inhibition and transportation behavior of novel β-enamino thiosemicarbazide derivatives by human serum albumin. J. Mol. Liq. 2018, 254, 280–290. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 1st ed.; Springer: New York, NY, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Alam, P.; Abdelhameed, A.S.; Rajpoot, R.K.; Khan, R.H. Interplay of multiple interaction forces: Binding of tyrosine kinase inhibitor nintedanib with human serum albumin. J. Photochem. Photobiol. B Biol. 2016, 157, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, Y.; Lv, C.; Wang, L.; Ou, J.; Wang, M.; Liu, S. Impact of halogen substituents on interactions between 2-phenyl-2,3-dihydroquinazolin-4(1H)-one derivatives and human serum albumin. Molecules 2012, 17, 2000–2014. [Google Scholar] [CrossRef] [PubMed]
- Chaves, O.A.; Amorim, A.P.O.; Castro, L.H.E.; Sant’Anna, C.M.R.; de Oliveira, M.C.C.; Cesarin-Sobrinho, D.; Netto-Ferreira, J.C.; Ferreira, A.B.B. Fluorescence and docking studies of the interaction between human serum albumin and pheophytin. Molecules 2015, 20, 19526–19539. [Google Scholar] [CrossRef] [PubMed]
- Montalti, M.; Credi, A.; Prodi, L.; Gandolfi, M.T. Handbook of Photochemistry, 3rd ed.; CRC Press: Boca Raton, FL, USA; Taylor & Francis: Didcot, UK, 2006; pp. 415–466. ISBN 9780824723774. [Google Scholar]
- Chaves, O.A.; Soares, B.A.; Maciel, M.A.M.; Sant’Anna, C.M.R.; Netto-Ferreira, J.C.; Cesarin-Sobrinho, D.; Ferreira, A.B.B. A study of the interaction between trans-dehydrocrotonin, a bioactive natural 19-nor-clerodane, and serum albumin. J. Braz. Chem. Soc. 2016, 27, 1858–1865. [Google Scholar]
- Molina-Bolívar, J.A.; Ruiz, C.C.; Galisteo-González, F.; Donnell, M.M.-O.; Parra, A. Simultaneous presence of dynamic and sphere action component in the fluorescence quenching of human serum albumin by diphthaloylmaslinic acid. J. Lumin. 2016, 178, 259–266. [Google Scholar] [CrossRef]
- Sun, H.; Liu, Y.; Li, M.; Han, S.; Yang, X.; Liu, R. Toxic effects of chrysoidine on human serum albumin: Isothermal titration calorimetry and spectroscopic investigations. Luminescence 2016, 31, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Chaves, O.A.; Mathew, B.; Joy, M.; Lohidakshan, K.K.; Marathakam, A.; Netto-Ferreira, J.C. Introduction of fluorinated environment on metformin. Evaluation of its serum-albumin interaction with molecular modeling studies. J. Mol. Liq. 2018, 260, 186–194. [Google Scholar] [CrossRef]
- Bi, S.; Zhao, T.; Zhou, H.; Wang, Y.; Li, Z. Probing the interactions of bromchlorbuterol-HCl and phenylethanolamine A with HSA by multi-spectroscopic and molecular docking technique. J. Chem. Thermodyn. 2016, 97, 113–121. [Google Scholar] [CrossRef]
- Turro, N.J. Modern Molecular Photochemistry, 1st ed.; University Science Books: Sausalito, CA, USA, 1991; ISBN 978-0-935702-7-12. [Google Scholar]
- Wang, Q.; Liu, X.; Su, M.; Shi, Z.; Sun, H. Study on the interaction characteristics of cefamandole with bovine serum albumin by spectroscopic technique. Spectrochim. Acta Mol. Biomol. 2015, 136, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Chaves, O.A.; Jesus, C.S.H.; Henriques, E.S.; Brito, R.M.M.; Serpa, C. In-situ ultra-fast heat deposition does not perturb serum albumin structure. Photochem. Photobiol. Sci. 2016, 15, 1524–1535. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.T.; Shamsi, H.; Khan, A.U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharm. 2014, 11, 1785–1797. [Google Scholar] [CrossRef] [PubMed]
- Chaves, O.A.; Mathew, B.; Cesarin-Sobrinho, D.; Lakshminarayanan, B.; Joy, M.; Mathew, G.E.; Suresh, J.; Netto-Ferreira, J.C. Spectroscopic, zeta potential and molecular docking analysis on the interaction between human serum albumin and halogenated thienyl chalcones. J. Mol. Liq. 2017, 242, 1018–1026. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, R.; Wang, Y.; Ma, J.; Li, K.; Li, Z. Biophysical study on the interaction of an anesthetic, vecuronium bromide with human serum albumin using spectroscopic and calorimetric methods. J. Photochem. Photobiol. B Biol. 2014, 140, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.N.; Liu, J.; Hu, Z.D.; Chen, X.G. Interaction of wogonin with bovine serum albumin. Bioorg. Med. Chem. 2005, 13, 4124–4129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-X.; Xiong, H.-X.; Li, L.-W. Investigation on the protein-binding properties of icotinib by spectroscopic and molecular modeling method. Spectrochim. Acta Mol. Biomol. Spectrosc. 2016, 161, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- De Barros, L.S.; Chaves, O.A.; Schaeffer, E.; Sant’Anna, C.M.R.; Ferreira, A.B.B.; Cesarin-Sobrinho, D.; da Silva, F.A.; Netto-Ferreira, J.C. Evaluating the interaction between di-fluorinated chalcones and plasmatic albumin. J. Fluor. Chem. 2016, 190, 81–88. [Google Scholar] [CrossRef]
- Vignesh, G.; Sugumar, K.; Arunachalam, S.; Vignesh, S.; James, R.A.; Arun, R.; Premkumar, K. Studies on the synthesis, characterization, human serum albumin binding and biological activity of single chain surfactant–cobalt(III) complexes. Luminescence 2016, 31, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Barakat, C.; Patra, D. Combining time-resolved fluorescence with synchronous fluorescence spectroscopy to study bovine serum albumin-curcumin complex during unfolding and refolding processes. Luminescence 2013, 28, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Hosainzadeh, A.; Gharanfoli, M.; Saberi, M.R.; Chamani, J.K. Probing the interaction of human serum albumin with bilirubin in the presence of aspirin by multi-spectroscopic, molecular modeling and zeta potential techniques: Insight on binary and ternary systems. J. Biomol. Struct. Dyn. 2012, 29, 1013–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, X.; Ding, S.; Lei, Q.; Fang, W. Unfolding of human serum albumin by gemini and single-chain surfactants: A comparative study. Colloids Surf. A 2016, 495, 30–38. [Google Scholar] [CrossRef]
- Rastegari, B.; Karbalaei-Heidari, H.R.; Yousefi, R.; Zeinali, S.; Nabavizadeh, M. Interaction of prodigiosin with HSA and β-Lg: Spectroscopic and molecular docking studies. Bioorg. Med. Chem. 2016, 24, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Zhou, X.; Chen, J.; Zhang, Y. Binding of bezafibrate to human serum albumin: Insight into the non-covalent interaction of an emerging contaminant with biomacromolecules. Molecules 2012, 17, 6821–6831. [Google Scholar] [CrossRef] [PubMed]
- Matei, I.; Hillebrand, M. Interaction of kaempferol with human serum albumin: A fluorescence and circular dichroism study. J. Pharm. Biomed. Anal. 2010, 51, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, G.; Baig, M.H.; Lee, E.J.; Cho, W.K.; Ma, J.Y.; Choi, I. Biophysical study on the interaction between eperisone hydrochloride and human serum albumin using spectroscopic, calorimetric, and molecular docking analysis. Mol. Pharm. 2017, 14, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar] [PubMed]
- Yue, Y.; Chen, X.; Qin, J.; Yao, X. Characterization of the mangiferin-human serum albumin complex by spectroscopic and molecular modeling approaches. J. Pharm. Biomed. Anal. 2009, 49, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Qin, Q.; Chang, M.; Li, S.; Shi, X.; Xu, G. Molecular interaction study of flavonoids with human serum albumin using native mass spectrometry and molecular modeling. Anal. Bioanal. Chem. 2018, 410, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Yeggoni, D.P.; Rachamallu, A.; Kallubai, M.; Subramanyam, R. Cytotoxicity and comparative binding mechanism of piperine with human serum albumin and α-1-acid glycoprotein. J. Biomol. Struct. Dyn. 2015, 33, 1336–1351. [Google Scholar] [CrossRef] [PubMed]
- Suresh, D.V.; Mahesha, H.G.; Rao, A.G.A.; Srinivasan, K. Binding of bioactive phytochemical piperine with human serum albumin: A spectrofluorometric study. Biopolymers 2007, 84, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Naveenraj, S.; Anandan, S. Binding of serum albumins with bioactive substances—Nanoparticles to drugs. J. Photochem. Photobiol. C Rev. 2013, 14, 53–71. [Google Scholar] [CrossRef]








| T (K) | KSV (M−1) | kq (M−1 s−1) | Ka (M−1) | ΔH° (kJ/mol) | ΔS° (kJ/molK) | ΔG° (kJ/mol) |
|---|---|---|---|---|---|---|
| 296 | (1.47 ± 0.04) × 104 | 2.54 × 1012 | (3.70 ± 0.26) × 103 | −20.6 | ||
| 303 | (1.45 ± 0.02) × 104 | 2.50 × 1012 | (1.08 ± 0.26) × 104 | 149 ± 20 | 0.573 ± 0.069 | −24.6 |
| 310 | (1.33 ± 0.02) × 104 | 2.30 × 1012 | (5.79 ± 0.26) × 104 | −28.6 |
| Sample | Ka (M−1) | Ka (M−1) | Ka (M−1) | Ka (M−1) |
|---|---|---|---|---|
| Without Site Marker | Presence Warfarin | Presence Ibuprofen | Presence Digitoxin | |
| RPF101 | (5.79 ± 0.26) × 104 | (1.63 ± 0.26) × 104 | (5.10 ± 0.26) × 104 | (4.55 ± 0.26) × 104 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaves, O.A.; Tavares, M.T.; Cunha, M.R.; Parise-Filho, R.; Sant’Anna, C.M.R.; Netto-Ferreira, J.C. Multi-Spectroscopic and Theoretical Analysis on the Interaction between Human Serum Albumin and a Capsaicin Derivative—RPF101. Biomolecules 2018, 8, 78. https://doi.org/10.3390/biom8030078
Chaves OA, Tavares MT, Cunha MR, Parise-Filho R, Sant’Anna CMR, Netto-Ferreira JC. Multi-Spectroscopic and Theoretical Analysis on the Interaction between Human Serum Albumin and a Capsaicin Derivative—RPF101. Biomolecules. 2018; 8(3):78. https://doi.org/10.3390/biom8030078
Chicago/Turabian StyleChaves, Otávio Augusto, Maurício Temotheo Tavares, Micael Rodrigues Cunha, Roberto Parise-Filho, Carlos Maurício R. Sant’Anna, and José Carlos Netto-Ferreira. 2018. "Multi-Spectroscopic and Theoretical Analysis on the Interaction between Human Serum Albumin and a Capsaicin Derivative—RPF101" Biomolecules 8, no. 3: 78. https://doi.org/10.3390/biom8030078