Genome-Wide Identification and Functional Characterization of the Phosphate Transporter Gene Family in Sorghum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Identification of Potential PHT Genes in Sorghum and Analysis of Their Chromosomal Localization
2.3. Phylogenetic Analysis
2.4. Gene Structure Analysis and Identification of Conserved Motifs
2.5. Protein Properties and Sequence Analysis
2.6. Subcellular Localization
2.7. Expression Analysis
3. Results
3.1. Genome-Wide Identification of PHT Family Genes in Sorghum
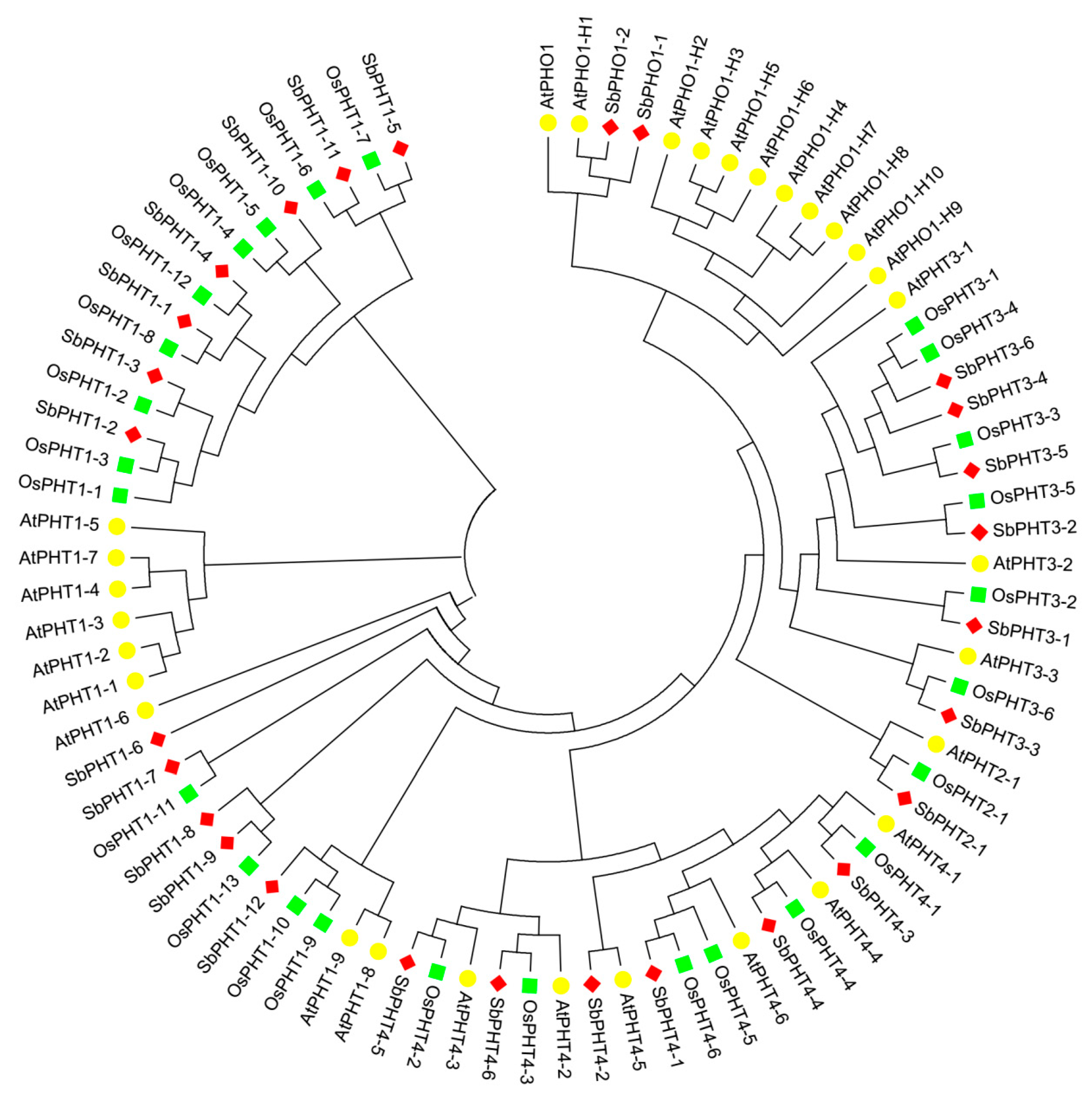
3.2. Phylogenetic Analysis of PHT Family Genes from Sorghum, Arabidopsis, and Rice
3.3. Genomic Organization and Protein Properties of Sorghum PHT Proteins
3.4. Gene Structure and Conserved Motifs of Sorghum PHT Proteins
3.5. Subcellular Localization and Transmembrane Domains of Sorghum PHT Proteins
3.6. Expression Profiles of Sorghum PHT Family Genes in Response to Low-Phosphate Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Plaxton, W.C.; Carswell, M.C. Metabolic aspects of the phosphate starvation response in plants. In Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization; Lerner, H.R., Ed.; M. Dekker: New York, NY, USA, 1999; pp. 350–372. [Google Scholar]
- Mimura, T. Regulation of phosphate transport and homeostasis in plant cells. Int. Rev. Cytol. 1999, 191, 149–200. [Google Scholar] [CrossRef]
- Lopez-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Muchhal, U.S.; Pardo, J.M.; Raghothama, K.G. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 10519–10523. [Google Scholar] [CrossRef] [PubMed]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef]
- Liu, T.-Y.; Huang, T.-K.; Tseng, C.-Y.; Lai, Y.-S.; Lin, S.-I.; Lin, W.-Y.; Chen, J.-W.; Chiou, T.-J. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 2012, 24, 2168–2183. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Pineros, M.; Wang, Z.; Wang, W.; Li, C.; Wu, Z.; Kochian, L.; Wu, P. Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 2012, 37, 1159–1170. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, X.; Zhang, L.; Lu, W.; Li, X.; Xiao, K. TaPht1;4, a high affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Funct. Plant Biol. 2013, 40, 329–341. [Google Scholar] [CrossRef]
- Rausch, C.; Daram, P.; Brunner, S.; Jansa, J.; Laloi, M.; Leggewie, G.; Amrhein, N.; Bucher, M. A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 2001, 414, 462–466. [Google Scholar] [CrossRef]
- Loth-Pereda, V.; Orsini, E.; Courty, P.-E.; Lota, F.; Kohler, A.; Diss, L.; Blaudez, D.; Chalot, M.; Nehls, U.; Bucher, M.; et al. Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiol. 2011, 156, 2141–2154. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, S.; Li, M.; Zhao, Z. Genomic identification and expression analysis of the phosphate transporter gene family in poplar. Front. Plant Sci. 2016, 7, 1398. [Google Scholar] [CrossRef]
- Wang, Y.; Ribot, C.; Rezzonico, E.; Poirier, Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 2004, 135, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Jin, Y.; Wussler, C.; Blancaflor, E.B.; Motes, C.M.; Versaw, W.K. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 2008, 77, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Teng, W.; Zhao, Y.-Y.; Zhao, X.-Q.; He, X.; Ma, W.-Y.; Deng, Y.; Chen, X.-P.; Tong, Y.-P. Genome-wide Identification, Characterization, and Expression Analysis of PHT1 Phosphate Transporters in Wheat. Front. Plant Sci. 2017, 8, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Chen, X.; Wang, H.; Liao, D.; Gu, M.; Qu, H.; Sun, S.; Xu, G. Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol. 2014, 14, 61. [Google Scholar] [CrossRef]
- Liu, F.; Chang, X.J.; Ye, Y.; Xie, W.B.; Wu, P.; Li, X.M. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, S.; Wu, X.; Wang, X.; Nan, Y.; Wang, D.; Chen, Q. Identification and characterization of phosphate transporter genes in potato. J. Biotechnol. 2017, 264, 17–28. [Google Scholar] [CrossRef]
- Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive Genomic Identification and Expression Analysis of the Phosphate Transporter (PHT) Gene Family in Apple. Front. Plant Sci. 2017, 8, 426. [Google Scholar] [CrossRef] [Green Version]
- Ai, P.; Sun, S.; Zhao, J.; Fan, X.; Xin, W.; Guo, Q.; Yu, L.; Shen, Q.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef]
- Qin, L.; Zhao, J.; Tian, J.; Chen, L.; Sun, Z.; Guo, Y.; Lu, X.; Gu, M.; Xu, G.; Liao, H. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 2012, 159, 1634–1643. [Google Scholar] [CrossRef]
- Karthikeyan, A.S.; Varadarajan, D.K.; Mukatira, U.T.; D’Urzo, M.P.; Damsz, B.; Raghothama, K.G. Regulated Expression of Arabidopsis Phosphate Transporters. Plant Physiol. 2002, 130, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Paszkowski, U.; Kroken, S.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, R.; Vasconcelos, M.J.V.; Zhao, S.; McElver, J.; Bruce, W.; Amrhein, N.; Raghothama, K.G.; Bucher, M. Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol. 2006, 8, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Wang, X.; Hu, R.; Wang, Y.; Xiao, C.; Jiang, Y.; Zhang, X.; Zheng, C.; Fu, Y.-F. The pattern of Phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biol. 2013, 3, 48. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Liu, J.; Dewbre, G.; Harrison, M.; Wang, Z.Y. Isolation and characterization of root-specific phosphate transporter promoters from Medicago truncatula. Plant Biol. 2006, 8, 439–449. [Google Scholar] [CrossRef]
- Liu, J.; Versaw, W.K.; Pumplin, N.; Gomez, S.K.; Blaylock, L.A.; Harrison, M.J. Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 2008, 283, 24673–24681. [Google Scholar] [CrossRef]
- Versaw, W.; Harrison, M. A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 2002, 14, 1751–1766. [Google Scholar] [CrossRef]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef]
- Nussaume, L.; Kanno, S.; Javot, H.; Marin, E.; Pochon, N.; Ayadi, A.; Nakanishi, T.M.; Thibaud, M.-C. Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Raghothama, K.G.; Smith, A.P. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Lapis-Gaza, H.R.; Jost, R.; Finnegan, P.M. Arabidopsis phosphate transporter 1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 2014, 14, 334. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Li, R.J.; Han, T.T.; Cai, W.; Fu, Z.W.; Lu, Y.T. Salt stress reduces root meristem size by nitric oxide mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shen, X.; Liang, H.; Wang, Y.; He, Z.; Zhang, D.; Chen, F. Isolation and Functional Analysis of PISTILLATA Homolog from Magnolia wufengensis. Front. Plant Sci. 2018, 9, 1743. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, C.; Shen, X.; Liang, H.; Wang, Y.; He, Z.; Zhang, D.; Chen, F. Comparative transcriptome analysis highlights the hormone effects on somatic embryogenesis in Catalpa bungei. Plant Reprod. 2019, 32, 141. [Google Scholar] [CrossRef]
- Ayadi, A.; David, P.; Arrighi, J.-F.; Chiarenza, S.; Thibaud, M.-C.; Nussaume, L.; Marín, E. Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 2015, 167, 1511–1526. [Google Scholar] [CrossRef]
- Dugas, D.V.; Monaco, M.K.; Olsen, A.; Klein, R.R.; Kumari, S.; Ware, D.; Klein, P.E. Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genom. 2011, 12, 514. [Google Scholar] [CrossRef]
- Yazawa, T.; Kawahigashi, H.; Matsumoto, T.; Mizuno, H. Simultaneous transcriptome analysis of Sorghum and Bipolaris sorghicola by using RNA-seq in combination with de novo transcriptome assembly. PLoS ONE 2013, 8, e62460. [Google Scholar] [CrossRef]
- Gelli, M.; Duo, Y.; Konda, A.R.; Zhang, C.; Holding, D.; Dweikat, I. Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genom. 2014, 15, 179. [Google Scholar] [CrossRef]
- Johnson, S.M.; Lim, F.L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Ramatoulaye, F.; Mady, C.; Fallou, S.; Amadou, K.; Cyril, D. Production and use of sorghum: A literature review. J. Nutr. Health Food Sci. 2016, 4, 1–4. [Google Scholar] [CrossRef]
- Anami, S.E.; Zhang, L.M.; Xia, Y.; Zhang, Y.M.; Liu, Z.Q.; Jing, H.C. Sweet sorghum ideotypes: Genetic improvement of the biofuel syndrome. Food Energy Secur. 2015, 4, 159–177. [Google Scholar] [CrossRef]
- Walder, F.; Brule, D.; Koegel, S.; Wiemken, A.; Boller, T.; Courty, P.E. Plant phosphorus acquisition in a common mycorrhizal network: Regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol. 2015, 205, 1632–1645. [Google Scholar] [CrossRef]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef]




| S. No. | Accession IDs | Gene IDs | Protein Length (aa) | Molecular Weight (kDa) | Isoelectric Point (pI) | Number of Introns | Number of Predicted Transmembrane Helices * | Predicted Subcellular Location ** |
|---|---|---|---|---|---|---|---|---|
| SbPHT1;1 | Sobic.001G234800 | Sb01g020570 | 541 | 58.78 | 7.62 | 0 | 11 | Plasma Membrane |
| SbPHT1;2 | Sobic.001G234900 | Sb01g020580 | 522 | 56.96 | 7.63 | 0 | 12 | Plasma Membrane |
| SbPHT1;3 | Sobic.001G502000 | Sb01g046890 | 522 | 57.14 | 8.7 | 0 | 11 | Plasma Membrane |
| SbPHT1;4 | Sobic.001G502100 | Sb01g046900 | 535 | 57.88 | 8.81 | 0 | 12 | Plasma Membrane |
| SbPHT1;5 | Sobic.001G513400 | Sb01g047910 | 541 | 58.25 | 6.46 | 0 | 11 | Plasma Membrane |
| SbPHT1;6 | Sobic.002G116100 | Sb02g009880 | 541 | 58.91 | 8.01 | 0 | 12 | Plasma Membrane |
| SbPHT1;7 | Sobic.003G243400 | Sb03g029970 | 554 | 60.26 | 7.61 | 1 | 12 | Plasma Membrane |
| SbPHT1;8 | Sobic.006G026800 | Sb06g002540 | 545 | 60.43 | 6.28 | 1 | 9 | — |
| SbPHT1;9 | Sobic.006G026900 | Sb06g002560 | 510 | 56.26 | 9.31 | 1 | 12 | Plasma Membrane |
| SbPHT1;10 | Sobic.006G027300 | Sb06g002800 | 536 | 58.63 | 6.8 | 1 | 10 | Plasma Membrane |
| SbPHT1;11 | Sobic.007G164400 | Sb07g023780 | 541 | 57.80 | 8.88 | 0 | 11 | — |
| SbPHT1;12 | Sobic.010G133300 | Sb10g012710 | 573 | 62.06 | 9.19 | 1 | 10 | Plasma Membrane |
| SbPHT2;1 | Sobic.004G199900 | Sb04g024630 | 572 | 59.31 | 9.48 | 2 | 13 | — |
| SbPHT3;1 | Sobic.001G428500 | Sb01g040260 | 371 | 39.73 | 8.97 | 5 | 0 | — |
| SbPHT3;2 | Sobic.002G224100 | Sb02g026490 | 351 | 37.31 | 9.33 | 5 | 0 | Chloroplast |
| SbPHT3;3 | Sobic.002G291800 | Sb02g032310 | 318 | 34.65 | 9.26 | 5 | 0 | — |
| SbPHT3;4 | Sobic.004G310300 | Sb04g034260 | 371 | 38.970 | 9.3 | 5 | 0 | — |
| SbPHT3;5 | Sobic.006G097400 | Sb06g018210 | 363 | 38.18 | 9.44 | 5 | 0 | — |
| SbPHT3;6 | Sobic.010G080900 | Sb10g007010 | 371 | 39.13 | 9.22 | 5 | 0 | — |
| SbPHT4;1 | Sobic.001G169100 | Sb01g014790 | 436 | 47.89 | 9.91 | 0 | 12 | Plasma Membrane |
| SbPHT4;2 | Sobic.002G293500 | Sb02g032440 | 542 | 58.07 | 5.85 | 14 | 9 | Plasma Membrane |
| SbPHT4;3 | Sobic.003G133700 | Sb03g011370 | 512 | 55.28 | 8.62 | 9 | 7 | — |
| SbPHT4;4 | Sobic.003G186800 | Sb03g025190 | 604 | 66.41 | 9.73 | 10 | 11 | Plasma Membrane |
| SbPHT4;5 | Sobic.003G358900 | Sb03g040080 | 517 | 55.66 | 9.92 | 7 | 10 | — |
| SbPHT4;6 | Sobic.009G157700 | Sb09g022090 | 535 | 56.17 | 9.76 | 7 | 11 | — |
| SbPHO1;1 | Sobic.003G101000 | Sb03g008460 | 851 | 95.45 | 9.04 | 10 | 5 | Plasma Membrane |
| SbPHO1;2 | Sobic.010G138800 | Sb10g014220 | 852 | 97.17 | 8.78 | 13 | 6 | — |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Yang, Y.; Liao, L.; Xu, J.; Liang, X.; Liu, W. Genome-Wide Identification and Functional Characterization of the Phosphate Transporter Gene Family in Sorghum. Biomolecules 2019, 9, 670. https://doi.org/10.3390/biom9110670
Wang J, Yang Y, Liao L, Xu J, Liang X, Liu W. Genome-Wide Identification and Functional Characterization of the Phosphate Transporter Gene Family in Sorghum. Biomolecules. 2019; 9(11):670. https://doi.org/10.3390/biom9110670
Chicago/Turabian StyleWang, Jiahui, Yang Yang, Lingzi Liao, Jiawei Xu, Xiao Liang, and Wen Liu. 2019. "Genome-Wide Identification and Functional Characterization of the Phosphate Transporter Gene Family in Sorghum" Biomolecules 9, no. 11: 670. https://doi.org/10.3390/biom9110670





