Chemical Constituents of the Leaves of Butterbur (Petasites japonicus) and Their Anti-Inflammatory Effects
Abstract
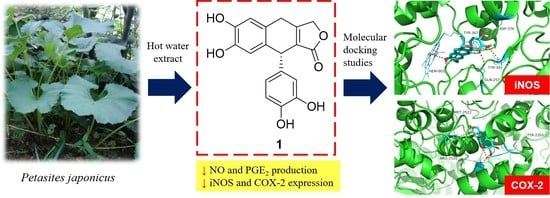
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Plant Material
2.3. Isolation of Compounds
2.3.1. Petasitesin A (1)
2.3.2. Petasitesin B (2)
2.4. Computational Methods
2.5. Measurement of NO Production
2.6. Measurement of PGE2
2.7. Measurement of iNOS and COX-2 Expression
2.8. Molecular Docking Studies
3. Results
3.1. Structure Elucidation of Compounds 1 and 2
3.2. Anti-inflammatory Effects of the Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kusano, A.; Seyama, Y.; Nagai, M.; Shibano, M.; Kusano, G. Effects of fukinolic acid and cimicifugic acids from Cimicifuga species on collagenolytic activity. Biol. Pharm. Bull. 2001, 24, 1198–1201. [Google Scholar] [CrossRef] [PubMed]
- Mizushina, Y.; Ishidoh, T.; Kamisuki, S.; Nakazawa, S.; Takemura, M.; Sugawara, F.; Yoshida, H.; Sakaguchi, K. Flavonoid glycoside: A new inhibitor of eukaryotic DNA polymerase α and a new carrier for inhibitor-affinity chromatography. Biochem. Biophys. Res. Commun. 2003, 301, 480–487. [Google Scholar] [CrossRef]
- Naya, K.; Hayashi, M.; Takagi, I.; Nakamura, S.; Kobayashi, M. The structural elucidation of sesquiterpene lactones from Petasites japonicus Maxim. Bull. Chem. Soc. Jpn. 1972, 45, 3673–3685. [Google Scholar] [CrossRef]
- Tori, M.; Kawahara, M.; Sono, M. Eremophilane-type sesquiterpenes from fresh rhizomes of Petasites japonicus. Phytochemistry 1997, 47, 401–409. [Google Scholar] [CrossRef]
- Wang, S.; Jin, D.Q.; Xie, C.; Wang, H.; Wang, M.; Xu, J.; Guo, Y. Isolation, characterization, and neuroprotective activities of sesquiterpenes from Petasites japonicus. Food Chem. 2013, 141, 2075–2082. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, H.; Tanaka, J.; Yamada, E.; Morikawa, T.; Kasajima, N.; Yoshikawa, M. Anti type I allergic property of Japanese butterbur extract and its mast cell degranulation inhibitory ingredients. J. Agric. Food Chem. 2006, 54, 2915–2920. [Google Scholar] [CrossRef] [PubMed]
- Sakamura, S.; Yoshihara, T.; Toyoda, K. The constituents of Petasites japonicus: Structures of fukiic acid and fukinolic acid. Agric. Biol. Chem. 1973, 37, 1915–1921. [Google Scholar] [CrossRef]
- Jeong, M.; Kim, H.M.; Kim, H.J.; Choi, J.H.; Jang, D.S. Kudsuphilactone B, a nortriterpenoid isolated from Schisandra chinensis fruit, induces caspase-dependent apoptosis in human ovarian cancer A2780 cells. Arch. Pharm. Res. 2017, 40, 500–508. [Google Scholar] [CrossRef]
- Du, Y.E.; Lee, J.S.; Kim, H.M.; Ahn, J.H.; Jung, I.H.; Ryu, J.H.; Choi, J.H.; Jang, D.S. Chemical constituents of the roots of Codonopsis lanceolata. Arch. Pharm. Res. 2018, 41, 1082–1091. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, H.; Ko, W.; Kim, D.C.; Kim, K.W.; Kwon, H.C.; Guo, Y.; Sohn, J.H.; Yim, J.H.; Kim, Y.C.; et al. Chemical constituents isolated from Antarctic marine-derived Aspergillus sp. SF-5976 and their anti-inflammatory effects in LPS-stimulated RAW 264.7 and BV2 cells. Tetrahedron 2017, 73, 3905–3912. [Google Scholar] [CrossRef]
- Ryu, S.M.; Lee, H.M.; Song, E.G.; Seo, Y.H.; Lee, J.; Guo, Y.; Kim, B.S.; Kim, J.J.; Hong, J.S.; Ryu, K.H.; et al. Antiviral activities of trichothecenes isolated from Trichoderma albolutescens against pepper mottle virus. J. Agric. Food. Chem. 2017, 65, 4273–4279. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.S.; Lee, K.G.; Lee, H.H.; Lee, H.J.; An, H.J.; Nam, J.H.; Jang, D.S.; Lee, K.T. α-Solanine Isolated from Solanum Tuberosum, L. cv Jayoung Abrogates LPS-Induced Inflammatory Responses Via NF-κB Inactivation in RAW 264.7 Macrophages and Endotoxin-Induced Shock Model in Mice. J. Cell. Biochem. 2016, 117, 2327–2339. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ren, Q.; Dong, B.; Shi, Z.; Zhang, J.; Jin, D.Q.; Xu, J.; Ohizumi, Y.; Lee, D.; Guo, Y. NO inhibitory constituents as potential anti-neuroinflammatory agents for AD from Blumea balsamifera. Bioorg. Chem. 2018, 76, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yang, X.; Ma, J.; Yang, Y.; Xie, C.; Tuerhong, M.; Jin, D.Q.; Xu, J.; Lee, D.; Ohizumi, Y.; et al. Nitric oxide inhibitory daphnane diterpenoids as potential anti-neuroinflammatory agents for AD from the twigs of Trigonostemon thyrsoideus. Bioorg. Chem. 2017, 75, 149–156. [Google Scholar] [CrossRef]
- Won, T.H.; Song, I.H.; Kim, K.H.; Yang, W.Y.; Lee, S.K.; Oh, D.C.; Oh, W.K.; Oh, K.B.; Shin, J. Bioactive metabolites from the fruits of Psoralea corylifolia. J. Nat. Prod. 2015, 78, 666–673. [Google Scholar] [CrossRef]
- Liu, Y.; Young, K.; Rakotondraibe, L.H.; Brodie, P.J.; Wiley, J.D.; Cassera, M.B.; Callmander, M.W.; Rakotondrajaona, R.; Rakotobe, E.; Rasamison, V.E.; et al. Antiproliferative compounds from Cleistanthus boivinianus from the Madagascar dry forest1. J. Nat. Prod. 2015, 78, 1543–1547. [Google Scholar] [CrossRef]
- Takahira, M.; Kusano, A.; Shibano, M.; Kusano, G.; Miyase, T. Piscidic acid and fukiic acid esters from Cimicifuga simplex. Phytochemistry 1998, 49, 2115–2119. [Google Scholar] [CrossRef]
- Wu, Q.Z.; Zhao, D.X.; Xiang, J.; Zhang, M.; Zhang, C.F.; Xu, X.H. Antitussive, expectorant, and anti-inflammatory activities of four caffeoylquinic acids isolated from Tussilago farfara. Pharm. Biol. 2016, 54, 1117–1124. [Google Scholar] [CrossRef]
- Park, H.Y.; Nam, M.H.; Lee, H.S.; Jun, W.; Hendrich, S.; Lee, K.W. Isolation of caffeic acid from Perilla frutescens and its role in enhancing γ-glutamylcysteine synthetase activity and glutathione level. Food Chem. 2010, 119, 724–730. [Google Scholar] [CrossRef]
- Noguchi, M.; Nagai, M.; Koeda, M.; Nakayama, S.; Sakurai, N.; Takahira, M.; Kusano, G. Vasoactive effects of cimicifugic acids C and D, and fukinolic acid in cimicifuga rhizome. Biol. Pharm. Bull. 1998, 21, 1163–1168. [Google Scholar] [CrossRef]
- Iwanaga, A.; Kusano, G.; Warashina, T.; Miyase, T. Hyaluronidase inhibitors from “Cimicifugae Rhizoma” (a mixture of the rhizomes of Cimicifuga dahurica and C. heracleifolia). J. Nat. Prod. 2010, 73, 573–578. [Google Scholar] [CrossRef] [PubMed]







| Position | 1 (in Acetone-d6) | 2 (in CD3OD) | ||
|---|---|---|---|---|
| δC | δH Multi (J in Hz) | δC | δH Multi (J in Hz) | |
| 1 | 129.8 | 127.0 | ||
| 2 | 136.9 | 130.1 | ||
| 3 | 116.9 | 6.60 s | 117.1 | 6.55 s |
| 4 | 145.1 | 145.5 | ||
| 5 | 145.3 | 145.7 | ||
| 6 | 115.7 | 6.72 s | 117.0 | 6.64 s |
| 7 | 29.0 | 3.86 d (23.0) | 39.6 | 2.83 d (14.5) |
| 3.62 overlapped | 2.67 d (14.5) | |||
| 8 | 160.9 | 78.0 | ||
| 9 | 72.4 | 4.97 d (17.0) | 80.1 | 4.17 d (10.0) |
| 4.89 d (17.0) | 3.93 d (10.0) | |||
| 1′ | 123.0 | 135.5 | ||
| 2′ | 116.2 | 6.59 d (2.0) | 116.0 | 6.58 d (2.0) |
| 3′ | 145.9 | 146.3 | ||
| 4′ | 144.6 | 144.8 | ||
| 5′ | 116.1 | 6.63 d (8.0) | 116.2 | 6.68 d (8.0) |
| 6′ | 120.3 | 6.45 dd (8.0, 2.0) | 120.2 | 6.52 dd (8.0, 2.0) |
| 7′ | 42.3 | 4.53 s | 47.4 | 4.18 d (3.0) |
| 8′ | 128.2 | 56.2 | 3.24 d (3.0) | |
| 9′ | 173.9 | 181.0 | ||
| Compound | IC50 (μM)a | |
|---|---|---|
| NO | PGE2 | |
| 1 | 15 ± 1.4 | 17 ± 3.2 |
| 2 | >50 | >50 |
| Cimicifugic acid D (3) | 12 ± 1.1 | 43 ± 7.9 |
| 4,5-Dicaffeoylquinic acid | 38.9 ± 0.72 | >50 |
| Caffeic acid | >50 | 45.7 ± 0.87 |
| Compound | −Log (FBE) | Targeting Residues | ||
|---|---|---|---|---|
| iNOS | COX-2 | iNOS | COX-2 | |
| 1 | −8.8 | −7.5 | ASP-376,TYR-367, TYR-341, GLN-257, HEM-901 | ARG-2120, TYR-3355, MET-2522 |
| 3 | −10.0 | −8.3 | ARG-260, ARG-375, ARG-382 TYR-341, TYR-367, TRP-340, GLN-257, HEM-901, VAL-346 | ARG-2120, TYR-2385, VAL-2116, SER-2530 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.S.; Jeong, M.; Park, S.; Ryu, S.M.; Lee, J.; Song, Z.; Guo, Y.; Choi, J.-H.; Lee, D.; Jang, D.S. Chemical Constituents of the Leaves of Butterbur (Petasites japonicus) and Their Anti-Inflammatory Effects. Biomolecules 2019, 9, 806. https://doi.org/10.3390/biom9120806
Lee JS, Jeong M, Park S, Ryu SM, Lee J, Song Z, Guo Y, Choi J-H, Lee D, Jang DS. Chemical Constituents of the Leaves of Butterbur (Petasites japonicus) and Their Anti-Inflammatory Effects. Biomolecules. 2019; 9(12):806. https://doi.org/10.3390/biom9120806
Chicago/Turabian StyleLee, Jin Su, Miran Jeong, Sangsu Park, Seung Mok Ryu, Jun Lee, Ziteng Song, Yuanqiang Guo, Jung-Hye Choi, Dongho Lee, and Dae Sik Jang. 2019. "Chemical Constituents of the Leaves of Butterbur (Petasites japonicus) and Their Anti-Inflammatory Effects" Biomolecules 9, no. 12: 806. https://doi.org/10.3390/biom9120806