Tomato Root Growth Inhibition by Salinity and Cadmium is Mediated by S-Nitrosative Modifications of ROS Metabolic Enzymes Controlled by S-Nitrosoglutathione Reductase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Plant Material
2.3. Root Sampling and Preparation of Root Extracts
2.4. Measurement of S-Nitrosothiol Content
2.5. Measurement of APX and GSNOR Activity
2.6. Measurement of NADPH Oxidase Activity
2.7. Quantification of Gene Expression by qPCR
2.8. Detection of NO, ONOO− and ROS
2.9. Purification and Detection of S-Nitrosated Proteins
2.10. Quantification of Protein Nitration
2.11. Image Analysis
2.12. Statistical Analysis
3. Results
3.1. Differences in Parameters of ROS and RNS Metabolism in the Roots of Solanum spp. Genotypes Grown Under Non-Stress Conditions
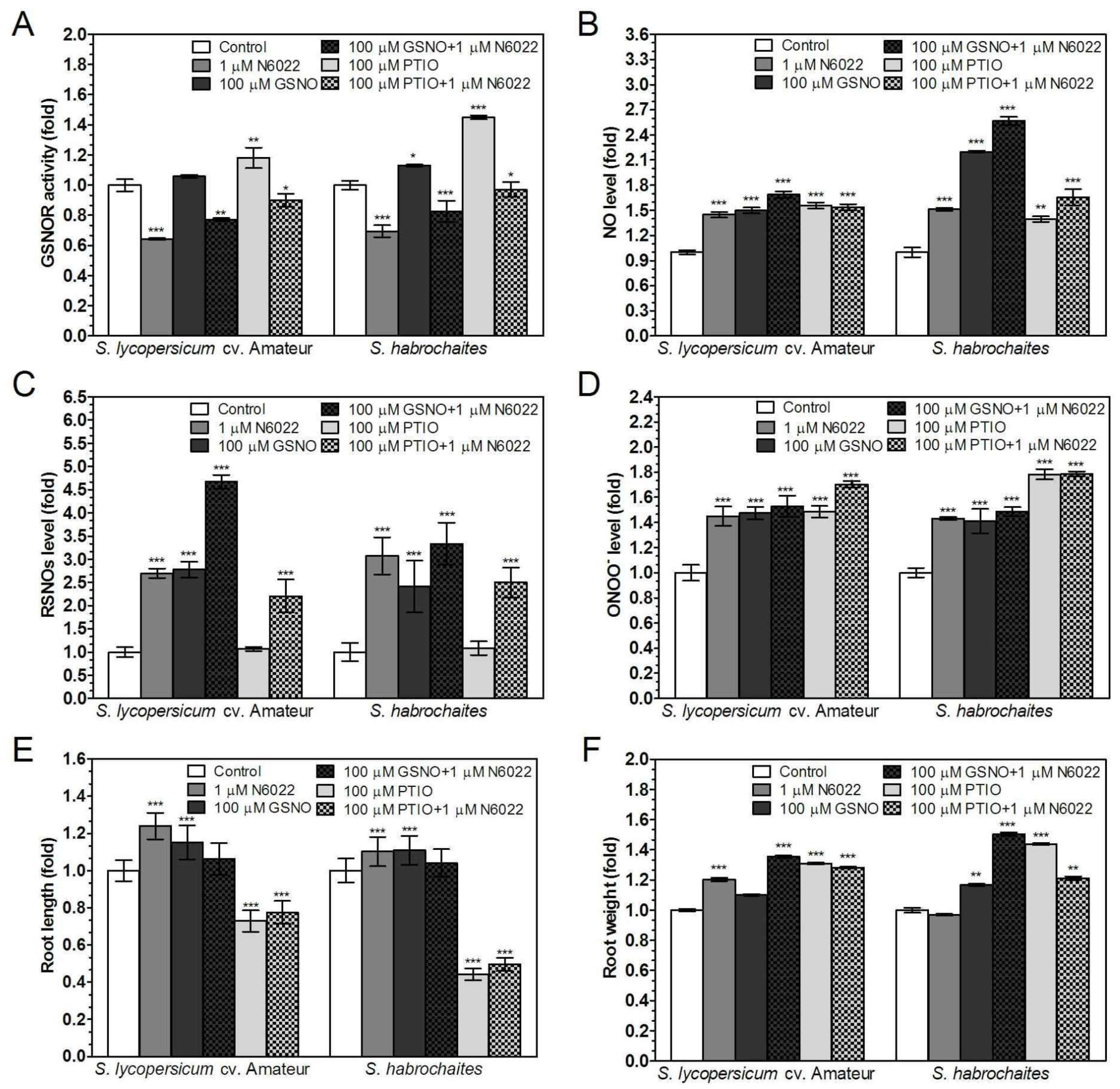
3.2. Inhibition of GSNOR Stimulated Root Growth in both Solanum spp. Genotypes
3.3. Effects of GSNOR Inhibition on Plant Root Growth are Mediated by Modulation of NO and S-Nitrosothiol Levels
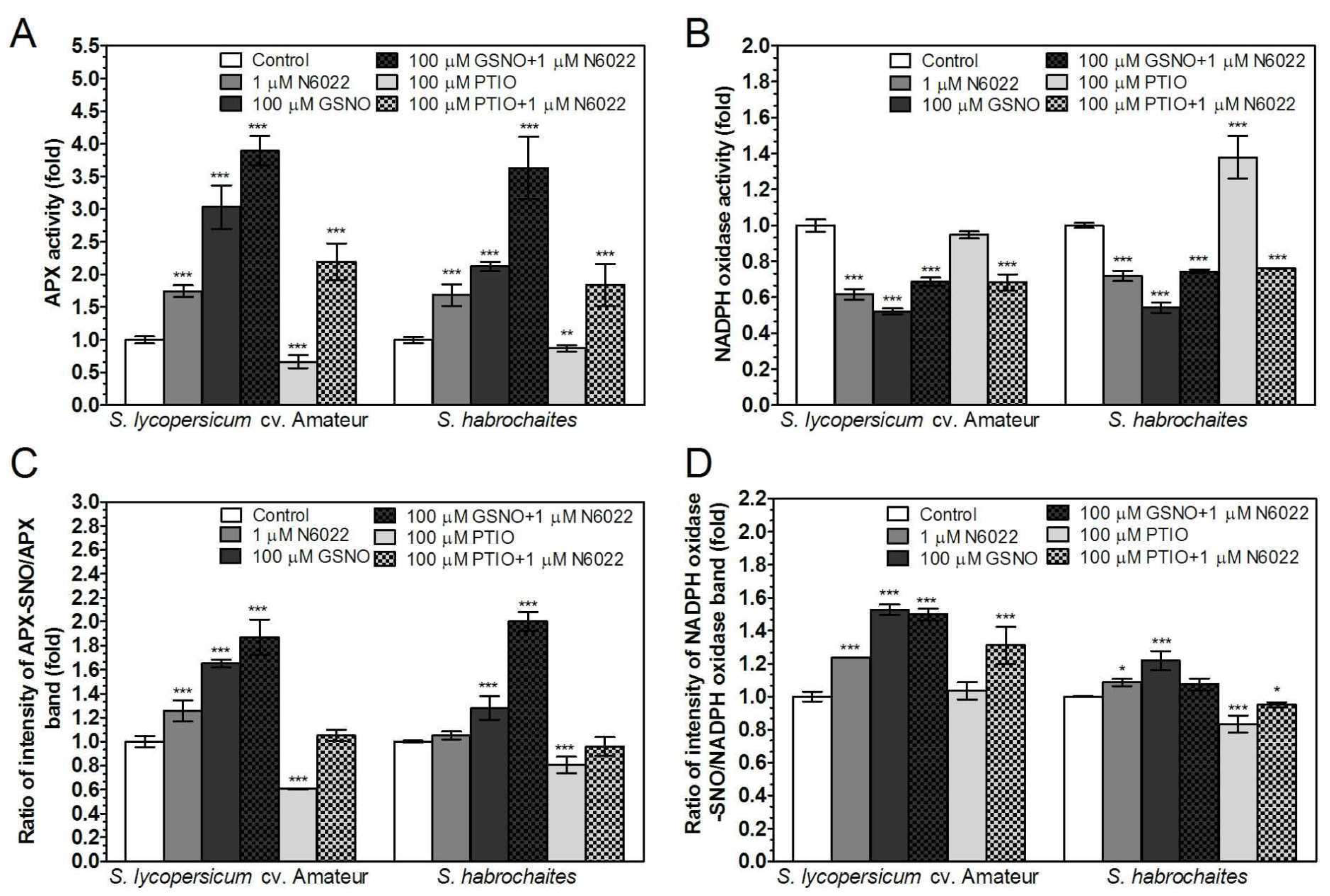
3.4. Activities of Enzymes of ROS Metabolism in the Roots Under Non-Stress Conditions are Regulated by GSNOR Through Protein S-Nitrosation
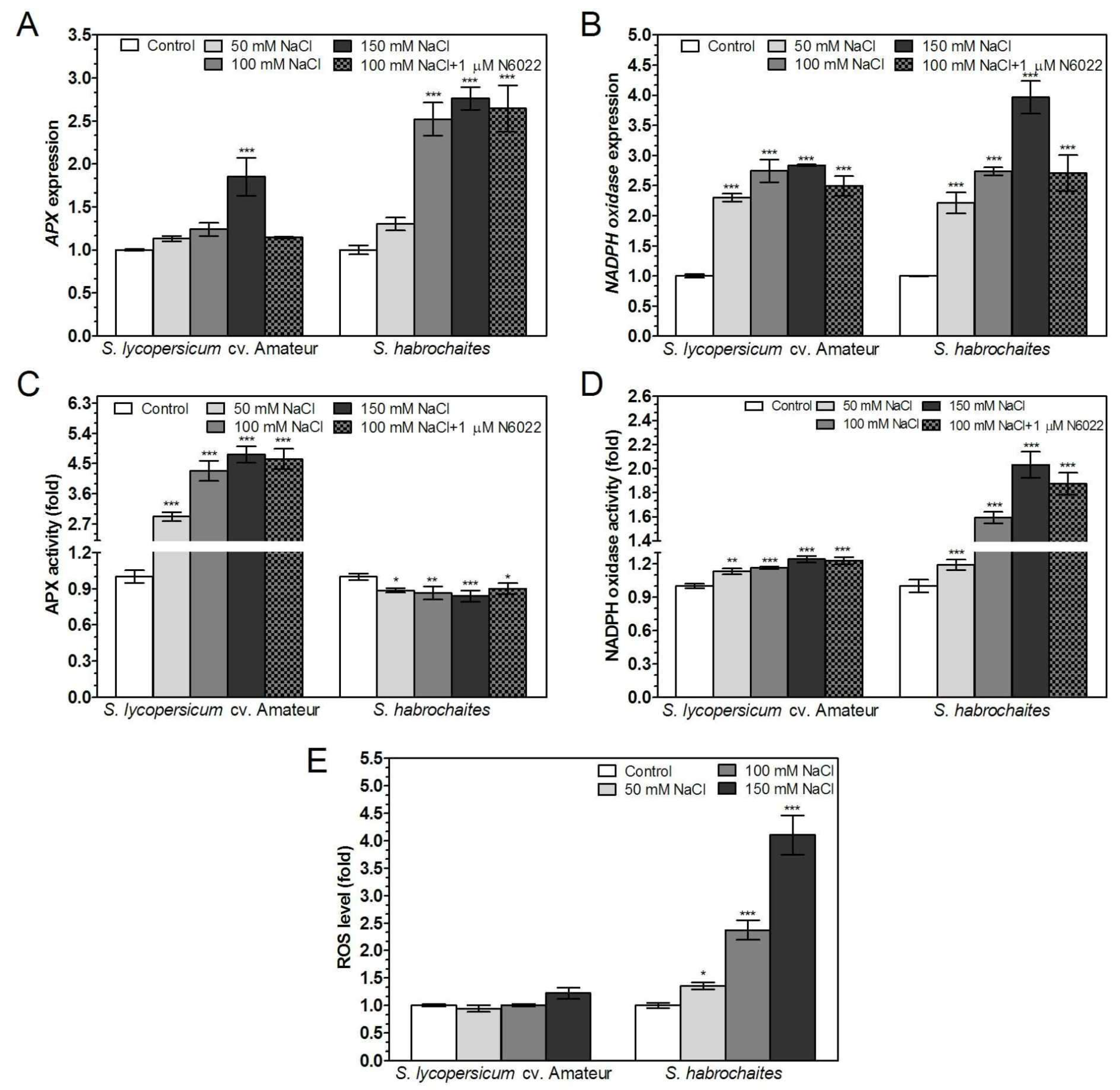
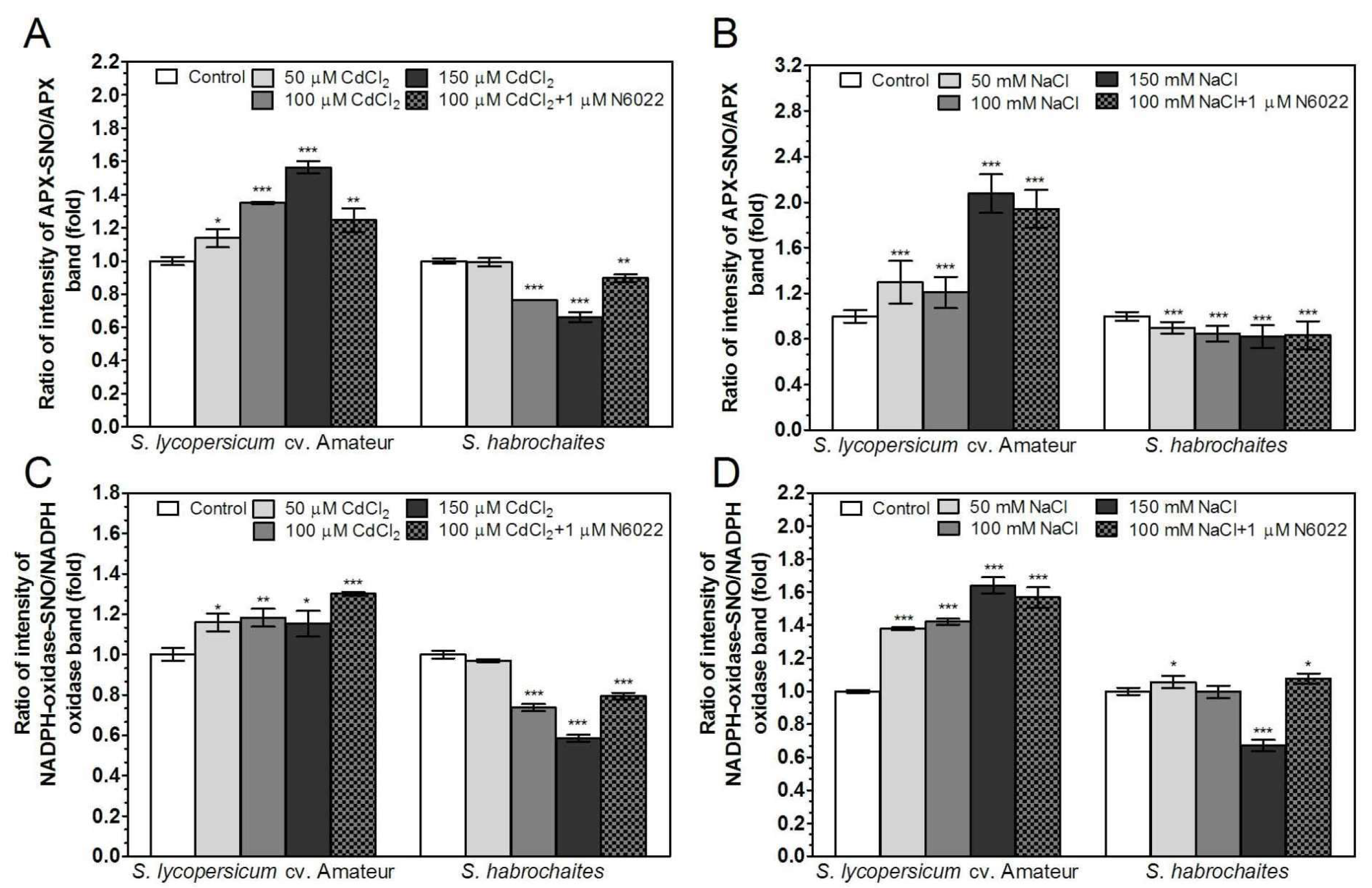
3.5. GSNOR is Differentially Involved in Root Responses of Solanum spp. Genotypes to Salinity and Heavy Metal Stress
3.6. APX and NADPH Oxidase Are Differentially Modulated by Abiotic Stress Conditions in Solanum spp. Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kolbert, Z. Implication of nitric oxide (NO) in excess element-induced morphogenic responses of the root system. Plant Physiol. Biochem. 2016, 101, 149–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.W.; Feechan, A.; Yin, M.; Saidi, N.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.G.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Groß, F.; Durner, J.; Gaupels, F. Nitric oxide, antioxidant and prooxidants in plant defence responses. Front. Plant Sci. 2013. [Google Scholar] [CrossRef]
- Beligni, M.V.; Lamattina, L. Nitric oxide counteracts cytotoxic processes mediated by reactive oxygen species in plant tissues. Planta 1999, 208, 337–344. [Google Scholar] [CrossRef]
- Fares, A.; Rossignol, M.; Peltier, J.B. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophy. Res. Comm. 2011, 416, 331–336. [Google Scholar] [CrossRef]
- Lin, A.; Wang, Y.; Tang, J.; Xue, P.; Li, C.; Liu, L.; Hu, B.; Yang, F.; Loake, G.J.; Chu, C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012, 158, 451–464. [Google Scholar] [CrossRef]
- Kato, H.; Takemoto, D.; Kawakita, K. Proteomic analysis of S-nitrosylated proteins in potato plant. Physiol. Plant. 2013, 148, 371–386. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; López-Jaramillo, J.; Padilla, M.N.; Carreras, A.; Corpas, F.J.; Barroso, J.B. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 2014, 65, 527–538. [Google Scholar] [CrossRef]
- Cheng, T.; Chen, J.; Abd Allah, E.F.; Wang, P.; Wang, G.; Hu, X.; Shi, J. Quantitative proteomics analysis reveals that S-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta 2015, 242, 1361–1390. [Google Scholar] [CrossRef]
- de Pinto, M.C.; Locato, V.; Sgobba, A.; Romero-Puertas, M.C.; Gadaleta, C.; Delledonne, M.; De Gara, L. S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco Bright Yellow-2 cells. Plant Physiol. 2013, 163, 1766–1775. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; Foresi, N.; Delledonne, M.; Lamattina, L. Auxin induces redox regulation of ascorbate peroxidase 1 activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis. J. Exp. Bot. 2013, 64, 3339–3349. [Google Scholar] [CrossRef]
- Yang, H.; Mu, J.; Chen, L.; Feng, J.; Hu, J.; Li, L. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 2015, 167, 1604–1615. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Mata-Pérez, C.; Valderrama, R.; Padilla, M.N.; López-Jaramillo, J.; Luque, F.; Corpas, F.J.; Barroso, J.B. Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S-nitrosylation. J. Exp. Bot. 2015, 66, 5983–5996. [Google Scholar] [CrossRef]
- Feechan, A.; Kwon, E.; Yun, B.W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef]
- Liu, L.; Hausladen, A.; Zeng, M.; Que, L.; Heitman, J.; Stamler, J.S. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 2001, 410, 490–494. [Google Scholar] [CrossRef]
- Lee, U.; Wie, C.; Fernandez, B.O.; Feelisch, M.; Vierling, E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 2008, 20, 786–802. [Google Scholar] [CrossRef]
- Leterrier, M.; Chaki, M.; Airaki, M.; Valderrama, R.; Palma, J.M.; Barroso, J.B.; Corpas, F.J. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 2011, 6, 789–793. [Google Scholar] [CrossRef] [Green Version]
- Airaki, M.; Leterrier, M.; Mateos, R.M.; Valderrama, R.; Chaki, M.; Barroso, J.B.; Del Río, L.A.; Palma, J.M.; Corpas, F.J. Metabolism of reactive oxygen species and reactive nitrogen species in pepper (Capsicum annuum L.) plants under low temperature stress. Plant Cell Environ. 2012, 35, 281–295. [Google Scholar] [CrossRef]
- Kwon, E.; Feechan, A.; Yun, B.W.; Hwang, B.H.; Pallas, J.A.; Kang, J.G.; Loake, G.J. AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 2012, 236, 887–900. [Google Scholar] [CrossRef]
- Kubienová, L.; Kopečný, D.; Tylichová, M.; Briozzo, P.; Skopalová, J.; Šebela, M.; Navrátil, M.; Tâche, R.; Luhová, L.; Barroso, J.B.; et al. Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum. Biochimie 2013, 95, 889–902. [Google Scholar] [CrossRef]
- Xu, S.; Guerra, D.; Lee, U.; Vierling, E. S-nitrosoglutathione reductases are low-copy number, cysteine-rich proteins in plants that control multiple developmental and defense responses in Arabidopsis. Front. Plant Sci. 2013. [Google Scholar] [CrossRef]
- Jahnová, J.; Luhová, L.; Petřivalský, M. S-Nitrosoglutathione Reductase-The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants 2019, 8, 48. [Google Scholar] [CrossRef]
- Camejo, D.; Romero-Puertas, M.C.; Rodríguez-Serrano, M.; Sandalio, L.M.; Lázaro, J.J.; Jiménez, A.; Sevilla, F. Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteom. 2013, 79, 87–99. [Google Scholar] [CrossRef]
- Gong, B.; Wen, D.; Wang, X.; Wei, M.; Yang, F.; Li, Y.; Shi, Q. S-nitrosoglutathione reductase-modulated redox signaling controls sodic alkaline stress responses in Solanum lycopersicum L. Plant Cell Physiol. 2015, 56, 790–802. [Google Scholar] [CrossRef]
- Yang, L.; Tian, D.; Todd, C.D.; Luo, Y.; Hu, X. Comparative proteome analyses reveal that nitric oxide is an important signal molecule in the response of rice to aluminum toxicity. J. Proteom. Res. 2013, 12, 1316–1330. [Google Scholar] [CrossRef]
- Frungillo, L.; Skelly, M.J.; Loake, G.J.; Spoel, S.H.; Salgado, I. S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat. Commun. 2014. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Dutta, S.; Mitra, M.; Agarwal, P.; Mahapatra, K.; De, S.; Sett, U.; Roy, S. Oxidative and genotoxic damages in plants in response to heavy metal stress and maintenance of genome stability. Plant Signal. Behav. 2018. [Google Scholar] [CrossRef]
- Mlíčková, K.; Luhová, L.; Lebeda, A.; Mieslerová, B.; Peč, P. Reactive oxygen species generation and peroxidase activity during Oidium neolycopersici infection on Lycopersicon species. Plant Physiol. Biochem. 2004, 42, 753–761. [Google Scholar] [CrossRef]
- Tománková, K.; Luhová, L.; Petřivalský, M.; Peč, P.; Lebeda, A. Biochemical aspects of reactive oxygen species formation in the interaction between Lycopersicon spp. and Oidium neolycopersici. Physiol Mol. Plant Pathol. 2006, 68, 22–32. [Google Scholar] [CrossRef]
- Piterková, J.; Hofman, J.; Mieslerová, B.; Sedlářová, M.; Luhová, L.; Lebeda, A.; Petřivalský, M. Dual role of nitric oxide in Solanum spp.-Oidium neolycopersici interactions. Environ. Exp. Bot. 2011, 74, 37–44. [Google Scholar] [CrossRef]
- Tichá, T.; Sedlářová, M.; Činčalová, L.; Trojanová, Z.D.; Mieslerová, B.; Lebeda, A.; Luhová, L.; Petřivalský, M. Involvement of S-nitrosothiols modulation by S-nitrosoglutathione reductase in defence responses of lettuce and wild Lactuca spp. to biotrophic mildews. Planta 2018, 247, 1203–1215. [Google Scholar] [CrossRef]
- Grandillo, S.; Chetelat, R.; Knapp, S.; Spooner, D.; Peralta, I.; Cammareri, M.; Ercolano, M.R. Solanum sect. Lycopersicon. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2011; pp. 129–215. [Google Scholar]
- Venema, J.H.; Dijk, B.E.; Bax, J.M.; van Hasselt, P.R.; Elzenga, J.T.M. Grafting tomato (Solanum lycopersicum) onto the rootstock of a high-altitude accession of Solanum habrochaites improves suboptimal-temperature tolerance. Environ. Exp. Bot. 2008, 63, 359–367. [Google Scholar] [CrossRef]
- Green, L.S.; Lawrence, E.C.; Patton, A.K.; Sun, X.; Rosenthal, G.J.; Richards, J.P. Mechanism of inhibition for N6022, a first-in-class drug targeting S-nitrosoglutathione reductase. Biochemistry 2012, 51, 2157–2168. [Google Scholar] [CrossRef]
- Moore, K.P.; Mani, A.R. Measurement of protein nitration and S-nitrosothiol formation in biology and medicine. Meth. Enzymol. 2002, 359, 256–268. [Google Scholar]
- Gow, A.; Doctor, A.; Mannick, J.; Gaston, B. S-nitrosothiol measurements in biological systems. J. Chromatogr. 2007, 851, 140–151. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for quantitation of mikrogram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kubienová, L.; Tichá, T.; Jahnová, J.; Luhová, L.; Mieslerová, B.; Petřivalský, M. Effect of abiotic stress stimuli on S nitrosoglutathione reductase in plants. Planta 2014, 239, 139–146. [Google Scholar] [CrossRef]
- Kaundal, A.; Rojas, C.M.; Mysore, K.S. Measurement of NADPH oxidase activity in plants. Bio-Protocol 2012. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001. [Google Scholar] [CrossRef]
- Lindermayr, C.; Saalbach, G.; Durner, J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef]
- Corpas, F.J.; Carreras, A.; Esteban, F.J.; Chaki, M.; Valderrama, R.; Del Río, L.A.; Barroso, J.B. Localization of S-nitrosothiols and assay of nitric oxide synthase and S-nitrosoglutathione reductase activity in plants. Meth. Enzymol. 2008, 437, 561–574. [Google Scholar]
- Correa-Aragunde, N.; Graziano, C.M.; Lamattina, L. Nitric oxide plays a central role in determining lateral root development in tomato. Planta 2004, 218, 900–905. [Google Scholar] [CrossRef]
- Pagnussat, G.C.; Simontacchi, M.; Puntarulo, S.; Lamattina, L. Nitric oxide is required for root organogenesis. Plant Physiol. 2002, 129, 954–956. [Google Scholar] [CrossRef]
- Fernández-Marcos, M.; Sanz, L.; Lewis, D.R.; Muday, G.K.; Lorenzo, O. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PINFORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. USA 2011, 108, 18506–18511. [Google Scholar] [CrossRef]
- Fernández-Marcos, M.; Sanz, L.; Lorenzo, O. Nitric oxide: An emerging regulator of cell elongation during primary root growth. Plant Signal. Behav. 2012, 7, 196–200. [Google Scholar] [CrossRef]
- Tichá, T.; Činčalová, L.; Kopečný, D.; Sedlářová, M.; Kopečná, M.; Luhová, L.; Petřivalský, M. Characterization of S-nitrosoglutathione reductase from Brassica and Lactuca spp. and its modulation during plant development. Nitric Oxide 2017, 68, 68–76. [Google Scholar] [CrossRef]
- Chen, R.; Sun, S.; Wang, C.; Li, Y.; Liang, Y.; An, F. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009, 19, 1377–1387. [Google Scholar] [CrossRef]
- Holzmeister, C.; Fröhlich, A.; Sarioglu, H.; Bauer, N.; Durner, J.; Lindermayr, C. Proteomic analysis of defense response of wildtype Arabidopsis thaliana and plants with impaired NO homeostasis. Proteomics 2011, 11, 1664–1683. [Google Scholar] [CrossRef]
- Shi, Y.F.; Wang, D.L.; Wang, C.; Culler, A.H.; Kreiser, M.A.; Suresh, J.; Cohen, J.D.; Pan, J.; Baker, B.; Liu, J.Z. Loss of GSNOR1 Function Leads to Compromised Auxin Signaling and Polar Auxin Transport. Mol. Plant 2015, 8, 1350–1365. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.Y.; Yu, X.; Cui, D.Y.; Sun, M.H.; Sun, W.N.; Tang, Z.C. The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 2007, 17, 638–649. [Google Scholar] [CrossRef]
- Zandonadi, D.B.; Santos, M.P.; Dobbss, L.B.; Olivares, F.L.; Canellas, L.P.; Binzel, M.L.; Okorokova-Facanha, A.L.; Facanha, A.R. Nitric oxide mediates humic acids-induced root development and plasma membrane H+-ATPase activation. Planta 2010, 231, 1026–1035. [Google Scholar] [CrossRef]
- Kopyra, M.; Gwozdz, E.A. Nitric oxide stimulates seed germination and counteracts the inhibitory effect of heavy metals and salinity on root growth of Lupinus luteus. Plant Physiol. Biochem. 2003, 41, 1011–1017. [Google Scholar] [CrossRef]
- Kong, J.; Dong, Y.; Song, Y.; Bai, X.; Tian, X.; Xu, L.; Liu, S.; He, Z. Role of exogenous nitric oxide in alleviating iron deficiency stress of peanut seedlings (Arachis hypogaea L.). J. Plant Growth Regul. 2016, 35, 31–43. [Google Scholar] [CrossRef]
- Piterková, J.; Luhová, L.; Hofman, J.; Turečková, V.; Novák, O.; Petřivalský, M.; Fellner, M. Nitric oxide is involved in light-specific responses of tomato during germination under normal and osmotic stress conditions. Ann. Bot. 2012, 110, 767–776. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, S.; Russo, A.; Samuni, A. Reactions of PTIO and carboxy-PTIO with ·NO, ·NO2, and O2−. J. Biol. Chem. 2003, 278, 50949–50955. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistence. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef]
- Wendehenne, D.; Durner, J.; Klessig, D.F. Nitric oxide: A new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 2004, 7, 449–455. [Google Scholar] [CrossRef]
- Tada, Y.; Mori, T.; Shinogi, T.; Yao, N.; Takahashi, S.; Betsuyaku, S.; Mayama, S. Nitric oxide and reactive oxygen species do not elicit hypersensitive cell death but induce apoptosis in the adjacent cells during the defense response of oat. Mol. Plant Microbe Inter. 2004, 17, 245–253. [Google Scholar] [CrossRef]
- Chaki, M.; Fernández-Ocana, A.M.; Valderrama, R.; Carreras, A.; Esteban, F.J.; Luque, F.; Gómez-Rodríguez, M.V.; Begara-Morales, J.C.; Corpas, F.J.; Barroso, J.B. Involvement of reactive nitrogen and oxygen species (RNS and ROS) in sunflower-mildew interaction. Plant Cell Physiol. 2009, 50, 265–279. [Google Scholar] [CrossRef]
- Delledonne, M.; Zeier, J.; Marocco, A.; Lamb, C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 2001, 98, 13454–13459. [Google Scholar] [CrossRef] [Green Version]
- Farnese, F.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When bad guys become good ones: The key role of reactive oxygen species and nitric oxide in the plant responses to abiotic stress. Front. Plant Sci. 2016. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trend. Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Clark, D.; Durner, J.; Navarre, D.A.; Klessig, D.F. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant Microbe Interact. 2000, 13, 1380–1384. [Google Scholar] [CrossRef]
- Keyster, M.; Klein, A.; Egbich, I.; Jacobs, A.; Ludidi, N. Nitric oxide increases the enzymatic activity of three ascorbate peroxidase isoforms in soybean root nodules. Plant Signal. Behav. 2011, 6, 956–961. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Yang, L.; Yang, Y.; Ahmad, P.; Yang, Y.; Hu, X. Deciphering the protective role of nitric oxide against salt stress at the physiological and proteomic levels in maize. J. Proteom. Res. 2012, 10, 4349–4364. [Google Scholar] [CrossRef]
- Martínez-Ruiz, A.; Lamas, S. Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: Convergences and divergences. Cardiovasc. Res. 2007, 75, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Kitajima, S. Hydrogen peroxide-mediated inactivation of two chloroplastic peroxidases, ascorbate peroxidase and 2-Cys peroxiredoxin. Photochem. Photobiol. 2008, 84, 1404–1409. [Google Scholar] [CrossRef]
- Gould, K.S.; Klinguer, A.; Pugin, A.; Wendehenne, D. Nitric oxide production in tobacco leaf cells: A generalized stress response? Plant Cell Environ. 2003, 26, 1851–1862. [Google Scholar] [CrossRef]
- Corpas, F.J.; Hayashi, M.; Mano, S.; Nishimura, M.; Barroso, J.B. Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiol. 2009, 151, 2083–2094. [Google Scholar] [CrossRef]
- David, A.; Yadav, S.; Bhatla, S.C. Sodium chloride stress induces nitric oxide accumulation in root tips and oil body surface accompanying slower oleosin degradation in sunflower seedlings. Physiol. Plant. 2010, 140, 342–354. [Google Scholar] [CrossRef]
- Valderrama, R.; Corpas, F.J.; Carreras, A.; Fernández-Ocaña, A.; Chaki, M.; Luque, F.; Gómez-Rodríguez, M.V.; Colmenero-Varea, P.; Del Río, L.A.; Barroso, J.B. Nitrosative stress in plants. FEBS Lett. 2007, 581, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Tanou, G.; Job, C.; Rajjou, L.; Arc, E.; Belghazi, M.; Diamantidis, G.; Job, D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 2009, 60, 795–804. [Google Scholar] [CrossRef]
- Tanou, G.; Filippou, P.; Belghazi, M.; Job, D.; Diamantidis, G.; Fotopoulos, V. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 2012, 72, 585–599. [Google Scholar] [CrossRef]
- Bandeoglu, E.; Eyidogan, F.; Yücel, M.; Öktem, H.A. Antioxidant responses of shoots and roots of lentil to NaCl-salinity stress. Plant Growth Regul. 2004, 42, 69–77. [Google Scholar] [CrossRef]
- Gueta-Dahan, Y.; Yaniv, Z.; Zilinskas, B.A.; Ben-Hayyim, G. Salt and oxidative stress: Similar and specific responses and their relation to salt tolerance in citrus. Planta 1997, 203, 460–469. [Google Scholar] [CrossRef]
- Molina, A.; Bueno, P.; Marín, M.C.; Rodríguez-Rosales, M.P.; Belver, A.; Venema, K.; Donaire, J.P. Involvement of endogenous salicylic acid content, lipoxygenase and antioxidant enzyme activities in the response of tomato cell suspension cultures to NaCl. New Phytol. 2002, 156, 409–415. [Google Scholar] [CrossRef]
- Manai, J.; Gouiab, H.; Corpas, F.J. Redox and nitric oxide homeostasis are affected in tomato (Solanum lycopersicum) roots under salinity-induced oxidative stress. J. Plant Physiol. 2014, 171, 1028–1035. [Google Scholar] [CrossRef]
- Kopyra, M.; Stachoń-Wilk, M.; Gwozdz, E.A. Effects of exogenous nitric oxide on the antioxidant capacity of cadmium-treated soybean cell suspension. Acta Physiol. Plant. 2006, 28, 525–536. [Google Scholar] [CrossRef]
- Arasimowicz-Jelonek, M.; Floryszak-Wieczorek, J.; Deckert, J.; Rucińska-Sobkowiak, R.; Gzyl, J.; Pawlak-Sprada, S.; Gwóźdź, E.A. Nitric oxide implication in cadmium-induced programmed cell death in roots and signaling response of yellow lupine plants. Plant Physiol. Biochem. 2012, 58, 124–134. [Google Scholar] [CrossRef]
- Piterková, J.; Luhová, L.; Navrátilová, B.; Sedlářová, M.; Petřivalský, M. Early and long-term responses of cucumber cells to high cadmium concentration are modulated by nitric oxide and reactive oxygen species. Acta Physiol. Plant. 2015. [Google Scholar] [CrossRef]
- Singh, H.P.; Batish, D.R.; Kaur, G.; Arora, K.; Kohli, R.K. Nitric oxide (as sodium nitroprusside) supplementation ameliorates Cd toxicity in hydroponically grown wheat roots. Environ. Exp. Bot. 2008, 63, 158–167. [Google Scholar] [CrossRef]
- Singh, H.P.; Kaur, S.; Batish, D.R.; Sharma, V.P.; Sharma, N.; Kohli, R.K. Nitric oxide alleviates arsenic toxicity by reducing oxidative damage in the roots of Oryza sativa (rice). Nitric Oxide 2009, 20, 289–297. [Google Scholar] [CrossRef]
- Farnese, F.S.; Oliveira, J.A.; Paiva, E.; Menezes-Silva, P.E.; da Silva, A.A.; Campos, F.V.; Ribeiro, C. The Involvement of Nitric Oxide in Integration of Plant Physiological and Ultrastructural Adjustments in Response to Arsenic. Front. Plant Sci. 2017. [Google Scholar] [CrossRef]
- Ismail, G.S.M. Protective role of nitric oxide against arsenic-induced damages in germinating mung bean seeds. Acta Physiol. Plant. 2012, 34, 1303–1311. [Google Scholar] [CrossRef]
- Piterková, J.; Petřivalský, M.; Luhová, L.; Mieslerová, B.; Sedlářová, M.; Lebeda, A. Local and systemic production of nitric oxide in tomato responses to powdery mildew infection. Mol. Plant Pathol. 2009, 10, 501–513. [Google Scholar] [CrossRef]
- Ortega-Galisteo, A.P.; Rodríguez-Serrano, M.; Pazmiño, D.M.; Gupta, D.K.; Sandalio, L.M.; Romero-Puertas, M.C. S-nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: Changes under abiotic stress. J. Exp. Bot. 2012, 63, 2089–2103. [Google Scholar] [CrossRef]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Rodríguez-Serrano, M.; Esteban, F.J.; Fernández-Ocaña, A.; Chaki, M.; Romero-Puertas, M.C.; Valderrama, R.; Sandalio, L.M.; et al. Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. J. Exp. Bot. 2006, 57, 1785–1793. [Google Scholar] [CrossRef] [Green Version]
- Leterrier, M.; Airaki, M.; Palma, J.M.; Chaki, M.; Barroso, J.B.; Corpas, F.J. Arsenic triggers the nitric oxide (NO) and S-nitrosoglutathione (GSNO) metabolism in Arabidopsis. Environ. Pollut. 2012, 166, 136–143. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.A.; Romero-Puertas, M.C.; Zabalza, A.N.A.; Corpas, F.J.; Gómez, M.; Del Río, L.A.; Sandalio, L.M. Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ. 2006, 29, 1532–1544. [Google Scholar] [CrossRef]
- Corpas, F.J.; Del Río, L.A.; Barroso, J.B. Post-translational modifications mediated by reactive nitrogen species: Nitrosative stress responses or components of signal transduction pathways? Plant Signal. Behav. 2008, 3, 301–303. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jedelská, T.; Šmotková Kraiczová, V.; Berčíková, L.; Činčalová, L.; Luhová, L.; Petřivalský, M. Tomato Root Growth Inhibition by Salinity and Cadmium is Mediated by S-Nitrosative Modifications of ROS Metabolic Enzymes Controlled by S-Nitrosoglutathione Reductase. Biomolecules 2019, 9, 393. https://doi.org/10.3390/biom9090393
Jedelská T, Šmotková Kraiczová V, Berčíková L, Činčalová L, Luhová L, Petřivalský M. Tomato Root Growth Inhibition by Salinity and Cadmium is Mediated by S-Nitrosative Modifications of ROS Metabolic Enzymes Controlled by S-Nitrosoglutathione Reductase. Biomolecules. 2019; 9(9):393. https://doi.org/10.3390/biom9090393
Chicago/Turabian StyleJedelská, Tereza, Veronika Šmotková Kraiczová, Lucie Berčíková, Lucie Činčalová, Lenka Luhová, and Marek Petřivalský. 2019. "Tomato Root Growth Inhibition by Salinity and Cadmium is Mediated by S-Nitrosative Modifications of ROS Metabolic Enzymes Controlled by S-Nitrosoglutathione Reductase" Biomolecules 9, no. 9: 393. https://doi.org/10.3390/biom9090393
APA StyleJedelská, T., Šmotková Kraiczová, V., Berčíková, L., Činčalová, L., Luhová, L., & Petřivalský, M. (2019). Tomato Root Growth Inhibition by Salinity and Cadmium is Mediated by S-Nitrosative Modifications of ROS Metabolic Enzymes Controlled by S-Nitrosoglutathione Reductase. Biomolecules, 9(9), 393. https://doi.org/10.3390/biom9090393





