A Path Forward: Promoting Microbial-Based Methods in the Control of Invasive Plant Species
Abstract
1. Introduction
2. Research Directions
2.1. Improving Beneficial Native Plant Phytochemical Production
2.2. Reducing Competitiveness in Invasive Plants
2.3. Increasing Herbicide Tolerance in Native Plants
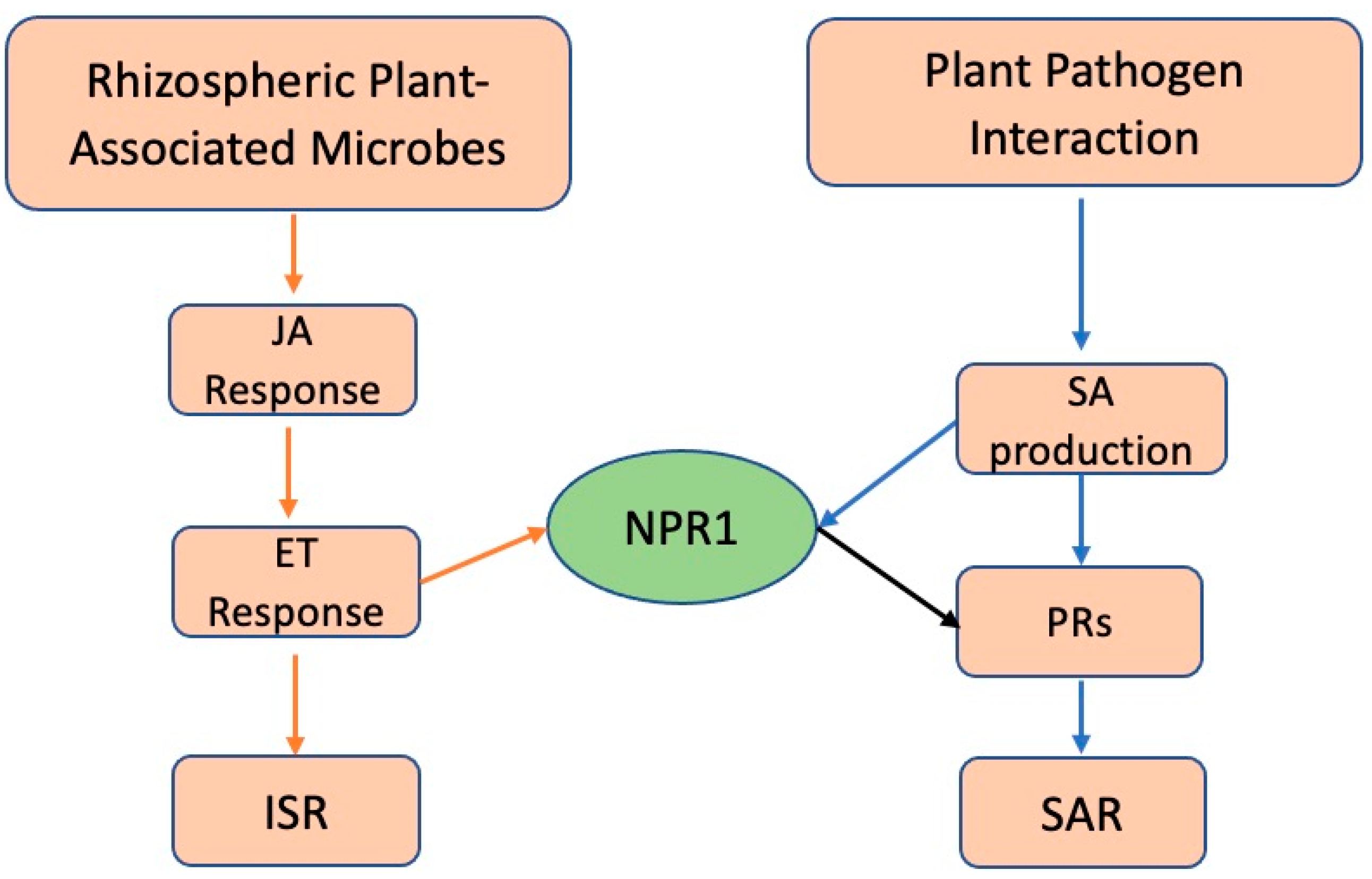
2.4. Facilitating Increased Pathogenicity in Invasive Plants
2.5. Concerns and Potential Problems with Microbial Deployment
3. Research Gaps, Future Directions, and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindow, S.E.; Leveau, J.H.J. Phyllosphere Microbiology. Curr. Opin. Biotechnol. 2002, 13, 238–243. [Google Scholar] [CrossRef]
- Bonkowski, M.; Villenave, C.; Griffiths, B. Rhizosphere Fauna: The Functional and Structural Diversity of Intimate Interactions of Soil Fauna with Plant Roots. Plant Soil 2009, 321, 213–233. [Google Scholar] [CrossRef]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil Origin and Plant Genotype Structure Distinct Microbiome Compartments in the Model Legume Medicago Truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef]
- Stone, J.K.; Bacon, C.W.; White, J.F., Jr. An overview of endophytic microbes: Endophytism defined. In Microbial Endophytes; Bacon, C.W., White, J.F., Jr., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 2000; pp. 3–30. [Google Scholar]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Goh, C.-H.; Veliz Vallejos, D.F.; Nicotra, A.B.; Mathesius, U. The Impact of Beneficial Plant-Associated Microbes on Plant Phenotypic Plasticity. J. Chem. Ecol. 2013, 39, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Garcia, B.J.; Furches, A.; Tuskan, G.A.; Jacobson, D. Plant Host-Associated Mechanisms for Microbial Selection. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research Priorities for Harnessing Plant Microbiomes in Sustainable Agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef]
- Kowalski, K.P.; Bacon, C.; Bickford, W.; Braun, H.; Clay, K.; Leduc-Lapierre, M.; Lillard, E.; McCormick, M.K.; Nelson, E.; Torres, M.; et al. Advancing the Science of Microbial Symbiosis to Support Invasive Species Management: A Case Study on Phragmites in the Great Lakes. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Weaver, M.A.; Boyette, C.D.; Hoagland, R.E. Management of Kudzu by the Bioherbicide, Myrothecium Verrucaria, Herbicides and Integrated Control Programmes. Biocontrol Sci. Technol. 2016, 26, 136–140. [Google Scholar] [CrossRef]
- Reichert Júnior, F.W.; Scariot, M.A.; Forte, C.T.; Pandolfi, L.; Dil, J.M.; Weirich, S.; Carezia, C.; Mulinari, J.; Mazutti, M.A.; Fongaro, G.; et al. New Perspectives for Weeds Control Using Autochthonous Fungi with Selective Bioherbicide Potential. Heliyon 2019, 5, e01676. [Google Scholar] [CrossRef]
- Lombard, L.; Houbraken, J.; Decock, C.; Samson, R.A.; Meijer, M.; Réblová, M.; Groenewald, J.Z.; Crous, P.W. Generic Hyper-Diversity in Stachybotriaceae. Persoonia 2016, 36, 156–246. [Google Scholar] [CrossRef]
- Clarke, T.C.; Shetty, K.G.; Jayachandran, K.; Norland, M.R. Myrothecium Verrucaria—A Potential Biological Control Agent for the Invasive ‘Old World Climbing Fern’ (Lygodium Microphyllum). BioControl 2007, 52, 399–411. [Google Scholar] [CrossRef][Green Version]
- Weaver, M.A.; Shearer, J.F.; Grodowitz, M.J.; Boyette, C.D. Potential of Myrothecium Species as Bioherbicides for Giant Salvinia (Salvinia Molesta). J. Aquat. Plant Manag. 2018, 56, 120–122. [Google Scholar]
- Harding, D.P.; Raizada, M.N. Controlling Weeds with Fungi, Bacteria and Viruses: A Review. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Shahrtash, M.; Brown, S.P. Drivers of Foliar Fungal Endophytic Communities of Kudzu (Pueraria Montana Var. Lobata) in the Southeast United States. Diversity 2020, 12, 185. [Google Scholar] [CrossRef]
- Lopez-Cervantes, J.; Thorpe, D.T. Microbial Composition Comprising Liquid Fertilizer and Processes for Agricultural Use. U.S. Patent Application 13/829,300, 3 October 2013. [Google Scholar]
- Stastny, M.; Sargent, R.D. Evidence for Rapid Evolutionary Change in an Invasive Plant in Response to Biological Control. J. Evol. Biol. 2017, 30, 1042–1052. [Google Scholar] [CrossRef]
- Brodeur, J. Host Specificity in Biological Control: Insights from Opportunistic Pathogens. Evol. Appl. 2012, 5, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.; Jumpponen, A.; Schlatter, D.C.; Paulitz, T.C.; Gardener, B.B.M.; Kinkel, L.L.; Garrett, K.A. Microbiome Networks: A Systems Framework for Identifying Candidate Microbial Assemblages for Disease Management. Phytopathology 2016, 106, 1083–1096. [Google Scholar] [CrossRef]
- Van der Heijden, M.G.A.; Hartmann, M. Networking in the Plant Microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef]
- Bonfante, P.; Anca, I.-A. Plants, Mycorrhizal Fungi, and Bacteria: A Network of Interactions. Annu. Rev. Microbiol. 2009, 63, 363–383. [Google Scholar] [CrossRef]
- Dames, J.F.; Ridsdale, C.J. What We Know about Arbuscular Mycorhizal Fungi and Associated Soil Bacteria. AJB 2012, 11, 13753–13760. [Google Scholar] [CrossRef]
- Keller, L.; Surette, M.G. Communication in Bacteria: An Ecological and Evolutionary Perspective. Nat. Rev. Microbiol. 2006, 4, 249–258. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Van, A.L.; Dufresne, A. The Importance of the Microbiome of the Plant Holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and Ecological Function of the Root Microbiome across Angiosperm Plant Species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.P.; Chen, L.; Schwier, M.; Koprivova, A.; Kopriva, S. Recent Advances in the Role of Plant Metabolites in Shaping the Root Microbiome. F1000Research 2020, 9. [Google Scholar] [CrossRef]
- Jones, D.L.; Nguyen, C.; Finlay, R.D. Carbon Flow in the Rhizosphere: Carbon Trading at the Soil–Root Interface. Plant Soil 2009, 321, 5–33. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Selvaraj, G.; Bais, H.P. Functional Soil Microbiome: Belowground Solutions to an Aboveground Problem. Plant Physiol. 2014, 166, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Abdiev, A.; Khaitov, B. Synergistic interactions among root-associated bacteria, rhizobia and chickpea under stress conditions. In Plant-Environment Interaction; Azzoz, M., Ahmad, P., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 250–262. ISBN 978-1-119-08100-5. [Google Scholar]
- Barret, M.; Morrissey, J.P.; O’Gara, F. Functional Genomics Analysis of Plant Growth-Promoting Rhizobacterial Traits Involved in Rhizosphere Competence. Biol. Fertil. Soils 2011, 47, 729. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Alford, É.R.; Perry, L.G.; Qin, B.; Vivanco, J.M.; Paschke, M.W. A Putative Allelopathic Agent of Russian Knapweed Occurs in Invaded Soils. Soil Biol. Biochem. 2007, 39, 1812–1815. [Google Scholar] [CrossRef]
- Li, Y.-P.; Feng, Y.-L.; Chen, Y.-J.; Tian, Y.-H. Soil Microbes Alleviate Allelopathy of Invasive Plants. Sci. Bull. 2015, 60, 1083–1091. [Google Scholar] [CrossRef]
- Hättenschwiler, S.; Vitousek, P.M. The Role of Polyphenols in Terrestrial Ecosystem Nutrient Cycling. Trends Ecol. Evol. 2000, 15, 238–243. [Google Scholar] [CrossRef]
- de Bruijn, W.J.C.; Gruppen, H.; Vincken, J.-P. Structure and Biosynthesis of Benzoxazinoids: Plant Defence Metabolites with Potential as Antimicrobial Scaffolds. Phytochemistry 2018, 155, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in Root Exudates of Maize Attract Pseudomonas Putida to the Rhizosphere. PLoS ONE 2012, 7, e35498. [Google Scholar] [CrossRef] [PubMed]
- Ditta, A.; Imtiaz, M.; Mehmood, S.; Rizwan, M.S.; Mubeen, F.; Aziz, O.; Qian, Z.; Ijaz, R.; Tu, S. Rock Phosphate-Enriched Organic Fertilizer with Phosphate-Solubilizing Microorganisms Improves Nodulation, Growth, and Yield of Legumes. Commun. Soil Sci. Plant Anal. 2018, 49, 2715–2725. [Google Scholar] [CrossRef]
- Ditta, A.; Muhammad, J.; Imtiaz, M.; Mehmood, S.; Qian, Z.; Tu, S. Application of Rock Phosphate Enriched Composts Increases Nodulation, Growth and Yield of Chickpea. Int. J. Recycl. Org. Waste Agricult. 2018, 7, 33–40. [Google Scholar] [CrossRef]
- Tiwari, G.; Duraivadivel, P.; Sharma, S.; Hariprasad, P. 1-Aminocyclopropane-1-Carboxylic Acid Deaminase Producing Beneficial Rhizobacteria Ameliorate the Biomass Characters of Panicum Maximum Jacq. by Mitigating Drought and Salt Stress. Sci. Rep. 2018, 8, 17513. [Google Scholar] [CrossRef] [PubMed]
- Sagar, A.; Sayyed, R.Z.; Ramteke, P.W.; Sharma, S.; Marraiki, N.; Elgorban, A.M.; Syed, A. ACC Deaminase and Antioxidant Enzymes Producing Halophilic Enterobacter Sp. PR14 Promotes the Growth of Rice and Millets under Salinity Stress. Physiol. Mol. Biol. Plant. 2020, 26, 1847–1854. [Google Scholar] [CrossRef]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant Growth Promoting Rhizobacteria (PGPR) as Green Bioinoculants: Recent Developments, Constraints, and Prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Theoharides, K.A.; Dukes, J.S. Plant Invasion across Space and Time: Factors Affecting Nonindigenous Species Success during Four Stages of Invasion. New Phytol. 2007, 256–273. [Google Scholar] [CrossRef] [PubMed]
- Callaway, R.M.; Ridenour, W.M. Novel Weapons: Invasive Success and the Evolution of Increased Competitive Ability. Front. Ecol. Environ. 2004, 2, 436–443. [Google Scholar] [CrossRef]
- Macel, M.; de Vos, R.C.H.; Jansen, J.J.; van der Putten, W.H.; van Dam, N.M. Novel Chemistry of Invasive Plants: Exotic Species Have More Unique Metabolomic Profiles than Native Congeners. Ecol. Evol. 2014, 4, 2777–2786. [Google Scholar] [CrossRef]
- Reinhart, K.O.; Callaway, R.M. Soil Biota and Invasive Plants. New Phytol. 2006, 170, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.; Reshi, Z.A.; Rasool, N. Plant Invasions Induce a Shift in Glomalean Spore Diversity. Trop. Ecol. 2010, 51, 317–323. [Google Scholar]
- Christina, M.; Rouifed, S.; Puijalon, S.; Vallier, F.; Meiffren, G.; Bellvert, F.; Piola, F. Allelopathic Effect of a Native Species on a Major Plant Invader in Europe. Sci. Nat. 2015, 102, 12. [Google Scholar] [CrossRef] [PubMed]
- Metlen, K.L.; Aschehoug, E.T.; Callaway, R.M. Competitive Outcomes between Two Exotic Invaders Are Modified by Direct and Indirect Effects of a Native Conifer. Oikos 2013, 122, 632–640. [Google Scholar] [CrossRef]
- Iason, G.R.; Lennon, J.J.; Pakeman, R.J.; Thoss, V.; Beaton, J.K.; Sim, D.A.; Elston, D.A. Does Chemical Composition of Individual Scots Pine Trees Determine the Biodiversity of Their Associated Ground Vegetation? Ecol. Lett. 2005, 8, 364–369. [Google Scholar] [CrossRef]
- Stinson, K.A.; Campbell, S.A.; Powell, J.R.; Wolfe, B.E.; Callaway, R.M.; Thelen, G.C.; Hallett, S.G.; Prati, D.; Klironomos, J.N. Invasive Plant Suppresses the Growth of Native Tree Seedlings by Disrupting Belowground Mutualisms. PLoS Biol. 2006, 4, e140. [Google Scholar] [CrossRef]
- Callaway, R.M.; Cipollini, D.; Barto, K.; Thelen, G.C.; Hallett, S.G.; Prati, D.; Stinson, K.; Klironomos, J. Novel Weapons: Invasive Plant Suppresses Fungal Mutualists in America but Not in Its Native Europe. Ecology 2008, 89, 1043–1055. [Google Scholar] [CrossRef]
- Lu-Irving, P.; Harenčár, J.G.; Sounart, H.; Welles, S.R.; Swope, S.M.; Baltrus, D.A.; Dlugosch, K.M. Native and Invading Yellow Starthistle (Centaurea Solstitialis) Microbiomes Differ in Composition and Diversity of Bacteria. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Mamet, S.D.; Redlick, E.; Brabant, M.; Lamb, E.G.; Helgason, B.L.; Stanley, K.; Siciliano, S.D. Structural Equation Modeling of a Winnowed Soil Microbiome Identifies How Invasive Plants Re-Structure Microbial Networks. ISME J. 2019, 13, 1988–1996. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Review: Endophytic Microbes and Their Potential Applications in Crop Management. Pest Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- MacDougall, A.S.; Gilbert, B.; Levine, J.M. Plant Invasions and the Niche. J. Ecol. 2009, 97, 609–615. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G. Effects of Exotic Plant Invasions on Soil Nutrient Cycling Processes. Ecosystems 2003, 6, 503–523. [Google Scholar] [CrossRef]
- Liao, C.; Peng, R.; Luo, Y.; Zhou, X.; Wu, X.; Fang, C.; Chen, J.; Li, B. Altered Ecosystem Carbon and Nitrogen Cycles by Plant Invasion: A Meta-Analysis. New Phytol. 2008, 177, 706–714. [Google Scholar] [CrossRef]
- Liao, Z.-Y.; Zhang, R.; Barclay, G.F.; Feng, Y.-L. Differences in Competitive Ability between Plants from Nonnative and Native Populations of a Tropical Invader Relates to Adaptive Responses in Abiotic and Biotic Environments. PLoS ONE 2013, 8, e71767. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, C.V.; Wren, I.F.; Herman, D.J.; Firestone, M.K. Plant Invasion Alters Nitrogen Cycling by Modifying the Soil Nitrifying Community. Ecol. Lett. 2005, 8, 976–985. [Google Scholar] [CrossRef]
- Lekberg, Y.; Gibbons, S.M.; Rosendahl, S.; Ramsey, P.W. Severe Plant Invasions Can Increase Mycorrhizal Fungal Abundance and Diversity. ISME J. 2013, 7, 1424–1433. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G.; Kourtev, P.; Huang, W. Changes in Soil Functions Following Invasions of Exotic Understory Plants in Deciduous Forests. Ecol. Appl. 2001, 11, 1287–1300. [Google Scholar] [CrossRef]
- McLeod, M.L.; Cleveland, C.C.; Lekberg, Y.; Maron, J.L.; Philippot, L.; Bru, D.; Callaway, R.M. Exotic Invasive Plants Increase Productivity, Abundance of Ammonia-Oxidizing Bacteria and Nitrogen Availability in Intermountain Grasslands. J. Ecol. 2016, 104, 994–1002. [Google Scholar] [CrossRef]
- Oses-Pedraza, R.; Torres-Díaz, C.; Lavín, P.; Retamales-Molina, P.; Atala, C.; Gallardo-Cerda, J.; Acuña-Rodríguez, I.S.; Molina-Montenegro, M.A. Root Endophytic Penicillium Promotes Growth of Antarctic Vascular Plants by Enhancing Nitrogen Mineralization. Extremophiles 2020, 24, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.M.; Lekberg, Y.; Mummey, D.L.; Sangwan, N.; Ramsey, P.W.; Gilbert, J.A. Invasive Plants Rapidly Reshape Soil Properties in a Grassland Ecosystem. mSystems 2017, 2. [Google Scholar] [CrossRef]
- Jack, C.N.; Petipas, R.H.; Cheeke, T.E.; Rowland, J.L.; Friesen, M.L. Microbial Inoculants: Silver Bullet or Microbial Jurassic Park? TIM 2020. [Google Scholar] [CrossRef]
- Kong, Z.; Hart, M.; Liu, H. Paving the Way From the Lab to the Field: Using Synthetic Microbial Consortia to Produce High-Quality Crops. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Zivanovic, A.; Rodgers, L. The Role of Fungal Endophytes in Plant Pathogen Resistance. Bios 2019, 89, 192–197. [Google Scholar] [CrossRef]
- Pant, P.; Pant, P. Ecological Restoration Techniques for Management of Degraded, Mined-Out Areas and the Role Played by Rhizospheric Microbial Communities. In Green Technologies and Environmental Sustainability; Singh, R., Kumar, S., Eds.; Springer: Cham, Switzerland, 2017; pp. 437–453. ISBN 978-3-319-50654-8. [Google Scholar]
- Domenech, J.; Reddy, M.S.; Kloepper, J.W.; Ramos, B.; Gutierrez-Mañero, J. Combined Application of the Biological Product LS213 with Bacillus, Pseudomonas or Chryseobacterium for Growth Promotion and Biological Control of Soil-Borne Diseases in Pepper and Tomato. Biocontrol 2006, 51, 245. [Google Scholar] [CrossRef]
- Rutherford, A.W.; Krieger-Liszkay, A. Herbicide-Induced Oxidative Stress in Photosystem II. Trends Biochem. Sci. 2001, 26, 648–653. [Google Scholar] [CrossRef]
- Demidchik, V. Mechanisms of Oxidative Stress in Plants: From Classical Chemistry to Cell Biology. Environ. Exp. Bot. 2015, 109, 212–228. [Google Scholar] [CrossRef]
- Singh, B.K.; Walker, A. Microbial Degradation of Organophosphorus Compounds. FEMS Microbiol. Rev. 2006, 30, 428–471. [Google Scholar] [CrossRef]
- Singh, B.; Singh, K. Microbial Degradation of Herbicides. Crit. Rev. Microbiol. 2016, 42, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xie, S. Isolation and Characterization of a High-Efficiency Soil Atrazine-Degrading Arthrobacter sp. Strain. Int. Biodeterior. Biodegrad. 2012, 71, 61–66. [Google Scholar] [CrossRef]
- Popov, V.H.; Cornish, P.S.; Sultana, K.; Morris, E.C.; Popov, V.H.; Cornish, P.S.; Sultana, K.; Morris, E.C. Atrazine Degradation in Soils: The Role of Microbial Communities, Atrazine Application History, and Soil Carbon. Soil Res. 2005, 43, 861–871. [Google Scholar] [CrossRef]
- Govantes, F.; Porrúa, O.; García-González, V.; Santero, E. Atrazine Biodegradation in the Lab and in the Field: Enzymatic Activities and Gene Regulation. Microb. Biotechnol. 2009, 2, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, C.S. Plant Protection and Rhizosphere Colonization of Barley by Seed Inoculated Herbicide Degrading Burkholderia (Pseudomonas) Cepacia DBO1(PRO101) in 2,4-D Contaminated Soil. Plant Soil 1997, 189, 139–144. [Google Scholar] [CrossRef]
- Zhang, L.; Hang, P.; Zhou, X.; Dai, C.; He, Z.; Jiang, J. Mineralization of the Herbicide Swep by a Two-Strain Consortium and Characterization of a New Amidase for Hydrolyzing Swep. Microb. Cell Fact. 2020, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Yan, X.; He, J.; Qiu, J.; Chen, Q. Comparative Genome Analysis Reveals the Evolution of Chloroacetanilide Herbicide Mineralization in Sphingomonas Wittichii DC-6. Arch. Microbiol. 2019, 201, 907–918. [Google Scholar] [CrossRef]
- Ahmad, K.S.; Gul, P.; Gul, M.M. Efficient Fungal and Bacterial Facilitated Remediation of Thiencarbazone Methyl in the Environment. Environ. Res. 2020, 188, 109811. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Elgorriaga, A.; Amrhein, N. Evidence for Two Distinct Phosphonate-Degrading Enzymes (C-P Lyases) in Arthrobacter Sp. GLP-1. Biodegradation 1991, 2, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Romeh, A. Biodegradation of Carbosulfan, Pirimicarb and Diniconazole Pesticides by Trichoderma Spp. J. Environ. Res. 2001, 3, 162–172. [Google Scholar]
- Harada, N.; Takagi, K.; Harazono, A.; Fujii, K.; Iwasaki, A. Isolation and Characterization of Microorganisms Capable of Hydrolysing the Herbicide Mefenacet. Soil Biol. Biochem. 2006, 38, 173–179. [Google Scholar] [CrossRef]
- Camacho-Morales, R.; Gerardo-Gerardo, J.; Guillén Navarro, K.; Sánchez, H. Ligninolytic Enzyme Production by White Rot Fungi during Paraquat (Herbicide) Degradation. Rev. Argent Microbiol. 2017, 49, 189–196. [Google Scholar] [CrossRef]
- Rønhede, S.; Jensen, B.; Rosendahl, S.; Kragelund, B.B.; Juhler, R.K.; Aamand, J. Hydroxylation of the Herbicide Isoproturon by Fungi Isolated from Agricultural Soil. Appl. Environ. Microbiol. 2005, 71, 7927–7932. [Google Scholar] [CrossRef]
- Carles, L.; Rossi, F.; Besse-Hoggan, P.; Blavignac, C.; Leremboure, M.; Artigas, J.; Batisson, I. Nicosulfuron Degradation by an Ascomycete Fungus Isolated From Submerged Alnus Leaf Litter. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Mougin, C.; Laugero, C.; Asther, M.; Dubroca, J.; Frasse, P.; Asther, M. Biotransformation of the Herbicide Atrazine by the White Rot Fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1994, 60, 705–708. [Google Scholar] [CrossRef]
- Cummins, I.; Cole, D.J.; Edwards, R. A Role for Glutathione Transferases Functioning as Glutathione Peroxidases in Resistance to Multiple Herbicides in Black-Grass. Plant J. 1999, 18, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Tétard-Jones, C.; Edwards, R. Potential Roles for Microbial Endophytes in Herbicide Tolerance in Plants. Pest Manag. Sci. 2016, 72, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Heil, M. Induced Systemic Resistance (ISR) against Pathogens—a Promising Field for Ecological Research. PPEES 2001, 4, 65–79. [Google Scholar] [CrossRef][Green Version]
- Ahsan, N.; Lee, D.-G.; Lee, K.-W.; Alam, I.; Lee, S.-H.; Bahk, J.D.; Lee, B.-H. Glyphosate-Induced Oxidative Stress in Rice Leaves Revealed by Proteomic Approach. Plant Physiol. Biochem. 2008, 46, 1062–1070. [Google Scholar] [CrossRef]
- Gao, Z.; Xie, W.; Ashraf, U.; Li, Y.; Ma, L.; Gui, R.; Pan, S.; Tian, H.; Duan, M.; Wang, S.; et al. Exogenous γ-Aminobutyric Acid (GABA) Application at Different Growth Stages Regulates 2-Acetyl-1-Pyrroline, Yield, Quality and Antioxidant Attributes in Fragrant Rice. J. Plant Interact. 2020, 15, 139–152. [Google Scholar] [CrossRef]
- Bailey, B.A.; Bae, H.; Strem, M.D.; Crozier, J.; Thomas, S.E.; Samuels, G.J.; Vinyard, B.T.; Holmes, K.A. Antibiosis, Mycoparasitism, and Colonization Success for Endophytic Trichoderma Isolates with Biological Control Potential in Theobroma Cacao. Biol. Control 2008, 46, 24–35. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Vogel, C.M.; Ueoka, R.; Schäfer, M.; Ryffel, F.; Müller, D.B.; Probst, S.; Kreuzer, M.; Piel, J.; Vorholt, J.A. Bipartite Interactions, Antibiotic Production and Biosynthetic Potential of the Arabidopsis Leaf Microbiome. Nat. Microbiol. 2018, 3, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Yang, N.; Wen, Z.; Sun, X.; Chai, Y.; Ma, Z. Wheat Microbiome Bacteria Can Reduce Virulence of a Plant Pathogenic Fungus by Altering Histone Acetylation. Nat. Commun. 2018, 9, 3429. [Google Scholar] [CrossRef]
- Parratt, S.R.; Laine, A.-L. The Role of Hyperparasitism in Microbial Pathogen Ecology and Evolution. ISME J. 2016, 10, 1815–1822. [Google Scholar] [CrossRef]
- Zelezniak, A.; Andrejev, S.; Ponomarova, O.; Mende, D.R.; Bork, P.; Patil, K.R. Metabolic Dependencies Drive Species Co-Occurrence in Diverse Microbial Communities. Proc. Natl. Acad. Sci. USA 2015, 112, 6449–6454. [Google Scholar] [CrossRef]
- Busby, P.E.; Ridout, M.; Newcombe, G. Fungal Endophytes: Modifiers of Plant Disease. Plant Mol. Biol. 2016, 90, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Van Wees, S.C.; De Swart, E.A.; Van Pelt, J.A.; Van Loon, L.C.; Pieterse, C.M. Enhancement of Induced Disease Resistance by Simultaneous Activation of Salicylate- and Jasmonate-Dependent Defense Pathways in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 8711–8716. [Google Scholar] [CrossRef]
- Pan, J.J.; May, G. Fungal-Fungal Associations Affect the Assembly of Endophyte Communities in Maize (Zea Mays). Microb. Ecol. 2009, 58, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Trapet, P.L.; Stringlis, I.A.; Kruijs, S.; Bakker, P.A.H.M.; Pieterse, C.M.J. Iron and Immunity. Annu. Rev. Phytopathol. 2017, 55, 355–375. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced Systemic Resistance (ISR) in Plants: Mechanism of Action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef]
- Newman, M.-A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (Microbe-Associated Molecular Pattern) Triggered Immunity in Plants. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Naidoo, S.; van den Berg, N. The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and Related Family: Mechanistic Insights in Plant Disease Resistance. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Nie, P.; Li, X.; Wang, S.; Guo, J.; Zhao, H.; Niu, D. Induced Systemic Resistance against Botrytis Cinerea by Bacillus Cereus AR156 through a JA/ET- and NPR1-Dependent Signaling Pathway and Activates PAMP-Triggered Immunity in Arabidopsis. Front. Plant Sci. 2017, 8, 238. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, H.; Behboudi, K.; Ahmadzadeh, M.; Javan-Nikkhah, M.; Zamioudis, C.; Pieterse, C.M.J.; Bakker, P.A.H.M. Induced Systemic Resistance in Cucumber and Arabidopsis Thaliana by the Combination of Trichoderma Harzianum Tr6 and Pseudomonas Sp. Ps14. Biol. Control 2013, 65, 14–23. [Google Scholar] [CrossRef]
- Ryu, C.-M.; Hu, C.-H.; Reddy, M.S.; Kloepper, J.W. Different Signaling Pathways of Induced Resistance by Rhizobacteria in Arabidopsis Thaliana against Two Pathovars of Pseudomonas Syringae. New Phytol. 2003, 160, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Kamle, M.; Borah, R.; Bora, H.; Jaiswal, A.K.; Singh, R.K.; Kumar, P. Systemic Acquired Resistance (SAR) and Induced Systemic Resistance (ISR): Role and Mechanism of Action Against Phytopathogens. In Fungal Biotechnology and Bioengineering; Hesham, A.E.-L., Upadhyay, R.S., Sharma, G.D., Manoharachary, C., Gupta, V.K., Eds.; Fungal Biology; Springer: Cham, Switzerland, 2020; pp. 457–470. ISBN 978-3-030-41870-0. [Google Scholar]
- de Boer, W.; Wagenaar, A.-M.; Klein Gunnewiek, P.J.A.; van Veen, J.A. In Vitro Suppression of Fungi Caused by Combinations of Apparently Non-Antagonistic Soil Bacteria. FEMS Microbiol. Ecol. 2007, 59, 177–185. [Google Scholar] [CrossRef]
- Venturi, V.; da Silva, D.P. Incoming Pathogens Team up with Harmless “resident” Bacteria. Trends Microbiol. 2012, 20, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Loon, L.C. van Salicylic Acid-Independent Plant Defence Pathways. Trends Plant Sci. 1999, 4, 52–58. [Google Scholar] [CrossRef]
- Paynter, Q.; Paterson, I.D.; Kwong, R.M. Predicting Non-Target Impacts. Curr. Opin. Insect Sci. 2020, 38, 79–83. [Google Scholar] [CrossRef]
- Molak, V. Fundamentals of Risk Analysis and Risk Management, 1st ed.; CRC Press: Boca Raton, FL, USA, 1996. [Google Scholar]
- Meena, S.S.; Mohanty, A. Ethical, Patent, and Regulatory Issues in Microbial Engineering. In Engineering of Microbial Biosynthetic Pathways; Singh, V., Singh, A.K., Bhargava, P., Joshi, M., Joshi, C.G., Eds.; Springer: Singapore, 2020; pp. 133–142. ISBN 9789811526046. [Google Scholar]
- Jamiołkowska, A. Natural Compounds as Elicitors of Plant Resistance Against Diseases and New Biocontrol Strategies. Agronomy 2020, 10, 173. [Google Scholar] [CrossRef]
- Thomloudi, E.-E.; Tsalgatidou, P.C.; Douka, D.; Spantidos, T.-N.; Dimou, M.; Venieraki, A.; Katinakis, P. Multistrain versus Single-Strain Plant Growth Promoting Microbial Inoculants—The Compatibility Issue. HPPJ. 2019, 12, 61–77. [Google Scholar] [CrossRef]
- Welsh, M.J.; Turner, J.A.; Epanchin-Niell, R.S.; Monge, J.J.; Soliman, T.; Robinson, A.P.; Kean, J.M.; Phillips, C.; Stringer, L.D.; Vereijssen, J.; et al. Approaches for Estimating Benefits and Costs of Interventions in Plant Biosecurity across Invasion Phases. Ecol. Appl. 2021, e2319. [Google Scholar] [CrossRef]
- Warziniack, T.; Haight, R.G.; Yemshanov, D.; Apriesnig, J.L.; Holmes, T.P.; Countryman, A.M.; Rothlisberger, J.D.; Haberland, C. Economics of Invasive Species. In Invasive Species in Forests and Rangelands of the United States; Poland, T.M., Patel-Weynand, T., Finch, D.M., Miniat, C.F., Hayes, D.C., Lopez, V.M., Eds.; Springer: Cham, Switzerland, 2021; pp. 305–320. ISBN 978-3-030-45366-4. [Google Scholar]
- Hanley, N.; Roberts, M. The Economic Benefits of Invasive Species Management. People Nat. 2019, 1, 124–137. [Google Scholar] [CrossRef]
- Mozelewski, T.G.; Scheller, R.M. Forecasting for Intended Consequences. Conserv. Sci. Pract. 2021, 3, e370. [Google Scholar] [CrossRef]
- Köhl, J.; Booij, K.; Kolnaar, R.; Ravensberg, W.J. Ecological Arguments to Reconsider Data Requirements Regarding the Environmental Fate of Microbial Biocontrol Agents in the Registration Procedure in the European Union. BioControl 2019, 64, 469–487. [Google Scholar] [CrossRef]
- Yang, Y.; Pratap Singh, R.; Song, D.; Chen, Q.; Zheng, X.; Zhang, C.; Zhang, M.; Li, Y. Synergistic Effect of Pseudomonas Putida II-2 and Achromobacter Sp. QC36 for the Effective Biodegradation of the Herbicide Quinclorac. Ecotoxicol. Environ. Saf. 2020, 188, 109826. [Google Scholar] [CrossRef]
- Zhang, L.; Hang, P.; Hu, Q.; Chen, X.-L.; Zhou, X.-Y.; Chen, K.; Jiang, J.-D. Degradation of Phenylurea Herbicides by a Novel Bacterial Consortium Containing Synergistically Catabolic Species and Functionally Complementary Hydrolases. J. Agric. Food Chem. 2018, 66, 12479–12489. [Google Scholar] [CrossRef] [PubMed]
- Pileggi, M.; Pileggi, S.A.V.; Sadowsky, M.J. Herbicide Bioremediation: From Strains to Bacterial Communities. Heliyon 2020, 6, e05767. [Google Scholar] [CrossRef]
- Villaverde, J.; Rubio-Bellido, M.; Lara-Moreno, A.; Merchan, F.; Morillo, E. Combined Use of Microbial Consortia Isolated from Different Agricultural Soils and Cyclodextrin as a Bioremediation Technique for Herbicide Contaminated Soils. Chemosphere 2018, 193, 118–125. [Google Scholar] [CrossRef]

| Species/Strain | Herbicide | Mode of Action | Citation |
|---|---|---|---|
| Bacteria | |||
| Pseudomonas sp. ADP. | Atrazine | Mineralization | [77] |
| Burkholderia (Pseudomonas) cepacia DBO1(pRO101) | 2,4-Dichlorophenoxyacetic acid | Biodegradation | [78] |
| Comamonas sp. SWP-3 | Swep | Hydrolysis | [79] |
| Alicycliphilus sp. PH-34 | Swep | Hydrolysis | [79] |
| Sphingomonas wittichii DC-6 | Chlorocetanilide | Mineralization | [80] |
| Pseudomonas syringae | Triazole | Biotransformation | [81] |
| Xanthomonas citri | Triazole | Biotransformation | [81] |
| Enterobacter cloacae K7 | Glyphosate | Biodegradation | [82] |
| Arthrobacter sp. GLP-1 | Glyphosate | Biodegradation | [82] |
| Fungi | |||
| Trichoderma viride | Pirimicarb | Biodegradation | [83] |
| Trichoderma harzianum | Pirimicarb | Biodegradation | [83] |
| Nocardioides sp. MFC-A | Mefenacet | Hydrolysis | [84] |
| Rhodococcus rhodochrous MFC-B | Mefenacet | Hydrolysis | [84] |
| Stenotrophomonas sp. | Mefenacet | Hydrolysis | [84] |
| Polyporus tricholoma | Paraquat | Enzymatic Degradation | [85] |
| Cilindrobasidium leave | Paraquat | Enzymatic Degradation | [85] |
| Deconica citrospora | Paraquat | Enzymatic Degradation | [85] |
| Aspergillus terrus | Triazole | Biotransformation | [81] |
| Penicillium chrysogenum | Triazole | Biotransformation | [81] |
| Mortierella sp. strain Gr4 | Isoproturon | Hydrolysis | [86] |
| Phoma cf. eupyrena Gr61 | Isoproturon | Hydrolysis | [86] |
| Alternaria sp. strain Gr174 | Isoproturon | Hydrolysis | [86] |
| Plectosphaerella cucumerina AR1 | Nicosulfuron | Hydrolysis | [87] |
| Phanerochaete chyrosporium | Atrazine | Biotransformation | [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahrtash, M.; Brown, S.P. A Path Forward: Promoting Microbial-Based Methods in the Control of Invasive Plant Species. Plants 2021, 10, 943. https://doi.org/10.3390/plants10050943
Shahrtash M, Brown SP. A Path Forward: Promoting Microbial-Based Methods in the Control of Invasive Plant Species. Plants. 2021; 10(5):943. https://doi.org/10.3390/plants10050943
Chicago/Turabian StyleShahrtash, Maryam, and Shawn P. Brown. 2021. "A Path Forward: Promoting Microbial-Based Methods in the Control of Invasive Plant Species" Plants 10, no. 5: 943. https://doi.org/10.3390/plants10050943
APA StyleShahrtash, M., & Brown, S. P. (2021). A Path Forward: Promoting Microbial-Based Methods in the Control of Invasive Plant Species. Plants, 10(5), 943. https://doi.org/10.3390/plants10050943






