Physiological and Metabolic Responses of Gac Leaf (Momordica cochinchinensis (Lour.) Spreng.) to Salinity Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Carbohydrate Analysis
2.3. Enzyme Activities
2.4. Chlorophyll Concentration
2.5. Electrolyte Leakage
2.6. Metabolite Profiling
2.7. Statistical Analysis and Data Analysis
3. Results
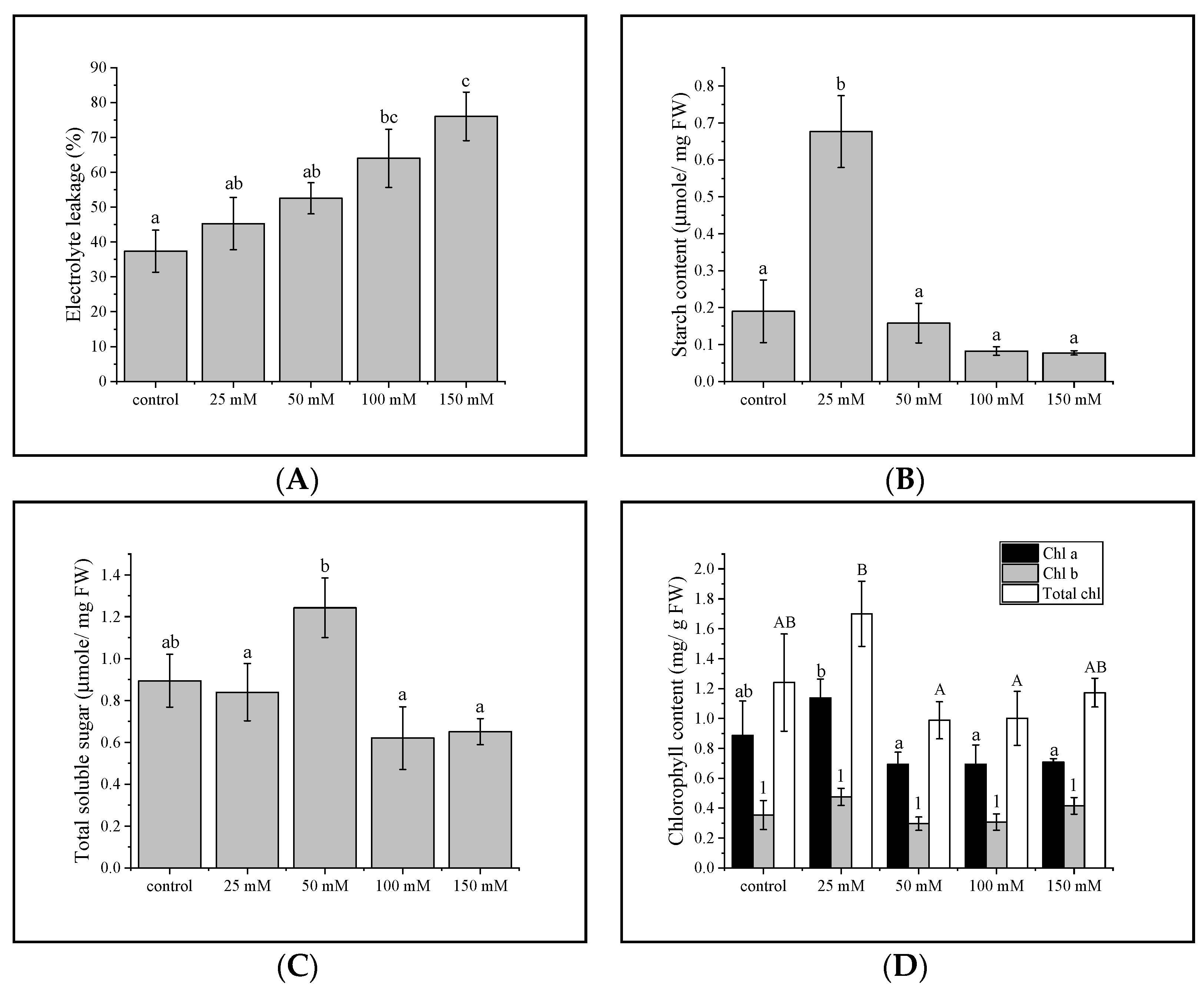
3.1. Electrolyte Leakage
3.2. Total Sugar and Starch Content
3.3. Chlorophyll Concentration in Gac Seedling Leaf
3.4. Antioxidant Enzyme Activities
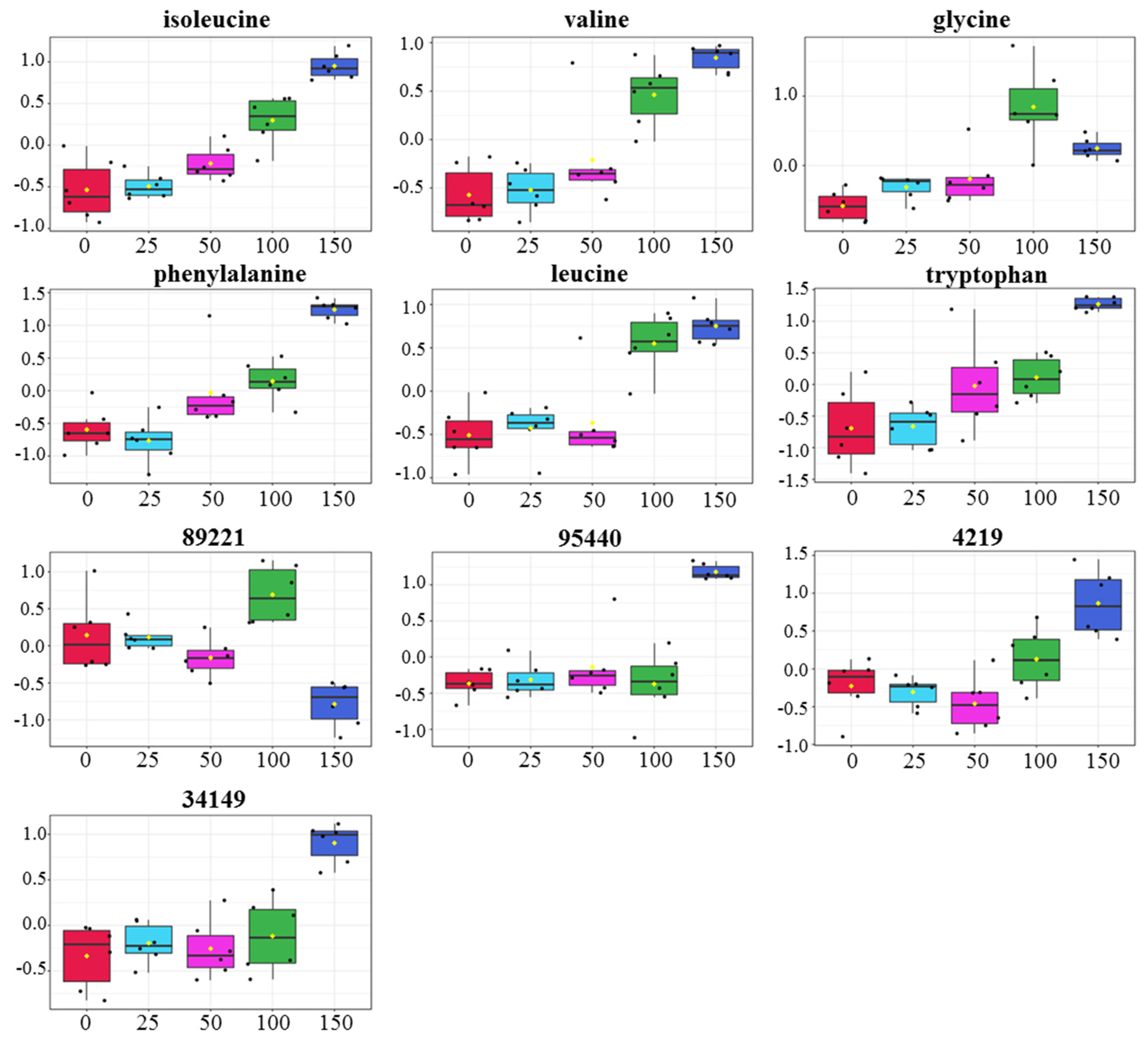
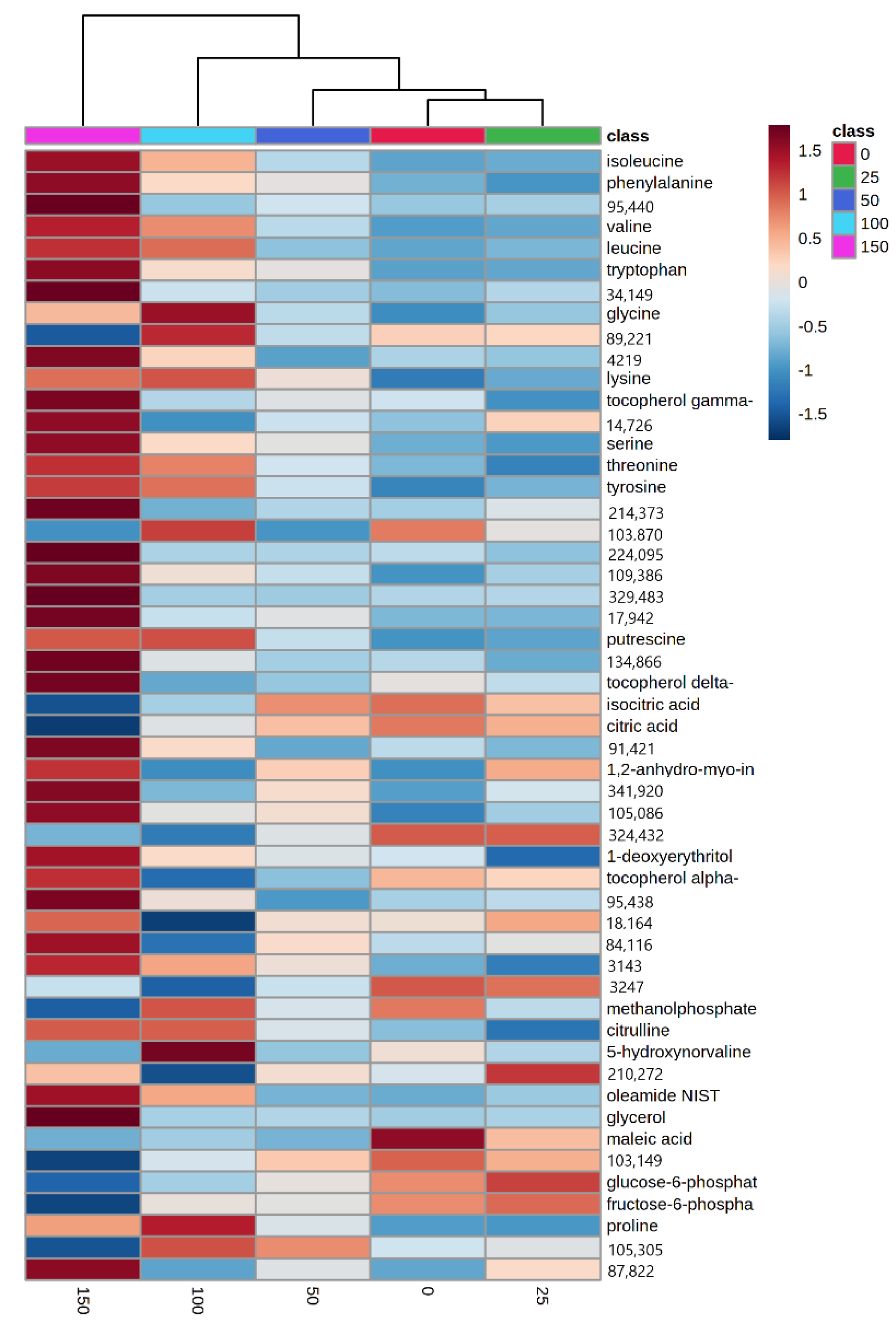
3.5. Relative Levels of Metabolites in Gac Seedling Leaf Concerning NaCl Concentration
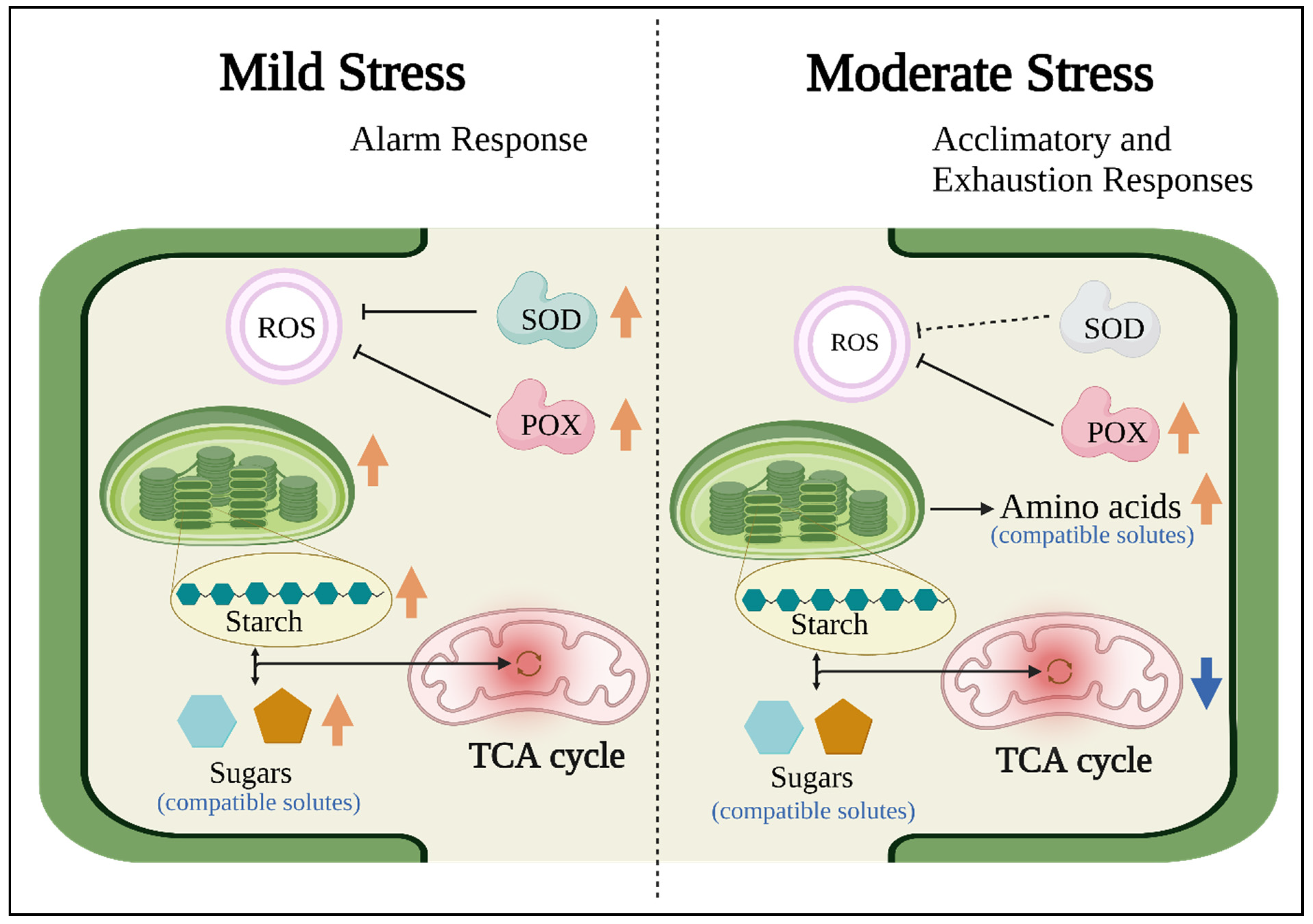
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ishida, B.K.; Turner, C.; Chapman, M.H.; McKeon, T.A. Fatty Acid and Carotenoid Composition of Gac (Momordica Cochinchinensis Spreng) Fruit. J. Agric. Food Chem. 2004, 52, 274–279. [Google Scholar] [CrossRef]
- Aoki, H.; Kieu, N.T.M.; Kuze, N.; Tomizaka, K.; Chuyen, N.V. Carotenoid Pigments in GAC Fruit ( Momordica Cochinchinensis SPRENG). Biosci. Biotechnol. Biochem. 2002, 66, 2479–2482. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Roh, H.-S.; Lee, S.; Jung, K.; Baek, K.-H.; Kim, K.H. Antiproliferative Effect of Momordica Cochinchinensis Seeds on Human Lung Cancer Cells and Isolation of the Major Constituents. Rev. Bras. Farmacogn. 2017, 27, 329–333. [Google Scholar] [CrossRef]
- Vuong, L.T.; Dueker, S.R.; Murphy, S.P. Plasma Beta-Carotene and Retinol Concentrations of Children Increase after a 30-d Supplementation with the Fruit Momordica Cochinchinensis (Gac). Am. J. Clin. Nutr. 2002, 75, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Osganian, S.K.; Stampfer, M.J.; Rimm, E.; Spiegelman, D.; Manson, J.E.; Willett, W.C. Dietary Carotenoids and Risk of Coronary Artery Disease in Women. Am. J. Clin. Nutr. 2003, 77, 1390–1399. [Google Scholar] [CrossRef]
- Nagarani, G.; Abirami, A.; Siddhuraju, P. Food Prospects and Nutraceutical Attributes of Momordica Species: A Potential Tropical Bioresources—A Review. Food Sci. Hum. Wellness 2014, 3, 117–126. [Google Scholar] [CrossRef]
- Mai, C.H.; Truong, V.; Frederic, D. Optimisation of Enzyme-Assisted Extraction of Oil Rich in Carotenoids from Gac Fruit (Momordica Cochinchinensis Spreng.). Food Technol. Biotechnol. 2013, 51, 488–499. [Google Scholar]
- Kubola, J.; Siriamornpun, S. Phytochemicals and Antioxidant Activity of Different Fruit Fractions (Peel, Pulp, Aril and Seed) of Thai Gac (Momordica Cochinchinensis Spreng). Food Chem. 2011, 127, 1138–1145. [Google Scholar] [CrossRef]
- Zhao, L.M.; Han, L.N.; Ren, F.Z.; Chen, S.H.; Liu, L.H.; Wang, M.X.; Sang, M.X.; Shan, B.E. An Ester Extract of Cochinchina Momordica Seeds Induces Differentiation of Melanoma B16 F1 Cells via MAPKs Signaling. Asian Pacific J. Cancer Prev. 2012, 13, 3795–3802. [Google Scholar] [CrossRef]
- Meng, L.-Y.; Liu, H.-R.; Shen, Y.; Yu, Y.-Q.; Tao, X. Cochinchina Momordica Seed Extract Induces G2/M Arrest and Apoptosis in Human Breast Cancer MDA-MB-231 Cells by Modulating the PI3K/Akt Pathway. Asian Pac. J. Cancer Prev. 2011, 12, 3483–3488. [Google Scholar]
- Liu, D.; Ford, K.L.; Roessner, U.; Natera, S.; Cassin, A.M.; Patterson, J.H.; Bacic, A. Rice Suspension Cultured Cells Are Evaluated as a Model System to Study Salt Responsive Networks in Plants Using a Combined Proteomic and Metabolomic Profiling Approach. Proteomics 2013, 13, 2046–2062. [Google Scholar] [CrossRef] [PubMed]
- Shannon, M.C.; Grieve, C.M. Tolerance of Vegetable Crops to Salinity. Sci. Hortic. 1998, 78, 5–38. [Google Scholar] [CrossRef]
- Bacilio, M.; Rodriguez, H.; Moreno, M.; Hernandez, J.-P.; Bashan, Y. Mitigation of Salt Stress in Wheat Seedlings by a Gfp-Tagged Azospirillum lipoferum. Biol. Fertil. Soils 2004, 40, 188–193. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of Glycine Betaine and Proline in Improving Plant Abiotic Stress Resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of Salinity and Water Content on Soil Microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- George, R.; McFarlane, D.; Nulsen, B. Salinity Threatens the Viability of Agriculture and Ecosystems in Western Australia. Hydrogeol. J. 1997, 5, 6–21. [Google Scholar] [CrossRef]
- Akbarimoghaddam, H.; Galavi, M.; Ghanbari, A.; Panjehkeh, N. Salinity Effects on Seed Germination and Seedling Growth of Bread Wheat Cultivars. Trakia J. Sci. 2011, 9, 43–50. [Google Scholar]
- Asada, K. Production and Scavenging of Reactive Oxygen Species in Chloroplasts and Their Functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Benzarti, M.; Rejeb, K.B.; Messedi, D.; Mna, A.B.; Hessini, K.; Ksontini, M.; Abdelly, C.; Debez, A. Effect of High Salinity on Atriplex portulacoides: Growth, Leaf Water Relations and Solute Accumulation in Relation with Osmotic Adjustment. South Afr. J. Bot. 2014, 95, 70–77. [Google Scholar] [CrossRef]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major Genes for Na+ Exclusion, Nax1 and Nax2 (Wheat HKT1;4 and HKT1;5), Decrease Na+ Accumulation in Bread Wheat Leaves under Saline and Waterlogged Conditions. J. Exp. Bot. 2011, 62, 2939–2947. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to Increasing the Salt Tolerance of Wheat and Other Cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [PubMed]
- Yasar, F.; Kusvuran, S.; Ellialtioglu, S. Determination of Anti-Oxidant Activities in Some Melon (Cucumis melo L.) Varieties and Cultivars under Salt Stress. J. Hortic. Sci. Biotechnol. 2006, 81, 627–630. [Google Scholar] [CrossRef]
- Rady, M.M.; Taha, R.S.; Mahdi, A.H.A. Proline Enhances Growth, Productivity and Anatomy of Two Varieties of Lupinus termis L. Grown under Salt Stress. South Afr. J. Bot. 2016, 102, 221–227. [Google Scholar] [CrossRef]
- Hadjigogos, K. The Role of Free Radicals in the Pathogenesis of Rheumatoid Arthritis. Panminerva Med. 2003, 45, 7–13. [Google Scholar]
- Yen, W.-J.; Chyau, C.-C.; Lee, C.-P.; Chu, H.-L.; Chang, L.-W.; Duh, P.-D. Cytoprotective Effect of White Tea against H2O2-Induced Oxidative Stress in Vitro. Food Chem. 2013, 141, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.M.; Chandrasekhar, T.; Hazara, M.; Sultan, Z.; Saleh, B.K.; Gopal, G.R. Recent Advances in Salt Stress Biology—A Review. Biotechnol. Mol. Biol. Rev. 2008, 3, 8–13. [Google Scholar]
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive Oxygen Species in Plants: Their Generation, Signal Transduction, and Scavenging Mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar]
- Erofeeva, E.A. Dependence of Guaiacol Peroxidase Activity and Lipid Peroxidation Rate in Drooping Birch (Betula Pendula Roth) and Tillet (Tilia Cordata Mill) Leaf on Motor Traffic Pollution Intensity. Dose-Response 2015, 13, 1559325815588510. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and Glutathione: Keeping Active Oxygen Under Control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Asada, K. The Water-Water Cycle in Chloroplasts: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Niyogi, K.K. Photoprotection Revisited: Genetic and Molecular Approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 333–359. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for Plant Stress Response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Fernie, A.R. The Use of Metabolomics to Dissect Plant Responses to Abiotic Stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a Tool to Investigate Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Munns, R.; Termaat, A. Whole-Plant Responses to Salinity. Aust. J. Plant Physiol. 1986, 13, 143. [Google Scholar] [CrossRef]
- Delauney, A.J.; Verma, D.P.S. Proline Biosynthesis and Osmoregulation in Plants. Plant J. 1993, 4, 215–223. [Google Scholar] [CrossRef]
- Flowers, T.J.; Flowers, S.A. Why Does Salinity Pose Such a Difficult Problem for Plant Breeders? Agric. Water Manag. 2005, 78, 15–24. [Google Scholar] [CrossRef]
- Paul, D. Osmotic Stress Adaptations in Rhizobacteria. J. Basic Microbiol. 2013, 53, 101–110. [Google Scholar] [CrossRef]
- Dasgan, H.Y.; Balacheva, E.; Yetişir, H.; Yarsi, G.; Altuntas, O.; Akhoundnejad, Y.; Coban, A. The Effectiveness of Grafting to Improve Salt Tolerance of Sensitive Melon When the Tolerant Melon Is Use as Rootstock. Procedia Environ. Sci. 2015, 29, 268. [Google Scholar] [CrossRef]
- Ford, C.W. Accumulation of Low Molecular Weight Solutes in Water-Stressed Tropical Legumes. Phytochemistry 1984, 23, 1007–1015. [Google Scholar] [CrossRef]
- Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Plants Use Calcium to Resolve Salt Stress. Trends Plant Sci. 1998, 3, 411–412. [Google Scholar] [CrossRef]
- Dubey, R.S.; Singh, A.K. Salinity Induces Accumulation of Soluble Sugars and Alters the Activity of Sugar Metabolising Enzymes in Rice Plants. Biologia Plant. 1999, 42, 233–239. [Google Scholar] [CrossRef]
- Flowers, T.J. Improving Crop Salt Tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Pattanagul, W.; Thitisaksakul, M. Effect of Salinity Stress on Growth and Carbohydrate Metabolism in Three Rice (Oryza Sativa L.) Cultivars Differing in Salinity Tolerance. Ind. J. Exp. Bio. 2008, 46, 736–742. [Google Scholar]
- Amirjani, M.R. Effect of Salinity Stress on Growth, Sugar Content, Pigments and Enzyme Activity of Rice. Int. J. Bot. 2011, 7, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.G.; Kobayashi, Y.; Sanuki, A.; Kondo, S.; Fukuda, N.; Ezura, H.; Sugaya, S.; Matsukura, C. Salinity Induces Carbohydrate Accumulation and Sugar-Regulated Starch Biosynthetic Genes in Tomato (Solanum lycopersicum L. Cv. ‘Micro-Tom’) Fruits in an ABA- and Osmotic Stress-Independent Manner. J. Exp. Bot. 2010, 61, 563–574. [Google Scholar] [CrossRef]
- Nemati, I.; Moradi, F.; Gholizadeh, S.; Esmaeili, M.A.; Bihamta, M.R. The Effect of Salinity Stress on Ions and Soluble Sugars Distribution in Leaves, Leaf Sheaths and Roots of Rice (Oryza Sativa L.) Seedlings. Plant Soil Environ. 2011, 57, 26–33. [Google Scholar] [CrossRef]
- Roberts, M.F. Organic Compatible Solutes of Halotolerant and Halophilic Microorganisms. Saline Syst. 2005, 1, 5. [Google Scholar] [CrossRef]
- Jumpa, T.; Pattanagul, W.; Songsri, P. Effects of Salinity Stress on Some Physiological Traits in Gac (Momordica Cochinchinensis (Lour.) Spreng.). Khon Kaen Agric. J. 2017, 45, 255–260. [Google Scholar]
- Robbins, N.S.; Pharr, D.M. Regulation of Photosynthetic Carbon Metabolism in Cucumber by Light Intencity and Photosynthetic Period. Plant Physiol. 1987, 85, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Madore, M.A. Carbohydrate Metabolism in Photosynthetic and Nonphotosynthetic Tissues of Variegated Leaves of Coleus blumei Benth. Plant Physiol. 1990, 93, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Su, W.; Li, H.; Guo, Z. Abscisic Acid Improves Drought Tolerance of Triploid ermudagrass and Involves H2O2- and NO-Induced Antioxidant Enzyme Activities. Plant Physiol. Biochem. 2009, 47, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sun, Z.; Henson, C.A. A Quantitative Assessment of the Importance of Barley Seed α-Amylase, β-Amylase, Debranching Enzyme, and α-Glucosidase in Starch Degradation. Arch. Biochem. Biophys. 1991, 284, 298–305. [Google Scholar] [CrossRef]
- Chandlee, J.M.; Scandalios, J.G. Analysis of Variants Affecting the Catalase Developmental Program in Maize Scutellum. Theor. Appl. Genet. 1984, 69, 71–77. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen Peroxide Is Scavenged by Ascorbate-Specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts Polyphenoloxidase in Beta vulgaris. Plant. Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Ling, Q.; Huang, W.; Jarvis, P. Use of a SPAD-502 Meter to Measure Leaf Chlorophyll Concentration in Arabidopsis thaliana. Photosynth. Res. 2011, 107, 209–214. [Google Scholar] [CrossRef]
- Yuan, Z.; Cao, Q.; Zhang, K.; Ata-Ul-Karim, S.T.; Tian, Y.; Zhu, Y.; Cao, W.; Liu, X. Optimal Leaf Positions for SPAD Meter Measurement in Rice. Front. Plant Sci. 2016, 7, 719. [Google Scholar] [CrossRef] [PubMed]
- Baninasab, B.; Ghobadi, C. Influence of Paclobutrazol and Application Methods on High-Temperature Stress Injury in Cucumber Seedlings. J. Plant Growth Regul. 2011, 30, 213–219. [Google Scholar] [CrossRef]
- Fiehn, O.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Lee, D.Y.; Lu, Y.; Moon, S.; Nikolau, B. Quality Control for Plant Metabolomics: Reporting MSI-Compliant Studies. Plant J. 2008, 53, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making Metabolomics More Meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards More Transparent and Integrative Metabolomics Analysis. Nucleic Acids Res. 2018, 46, 486–494. [Google Scholar] [CrossRef]
- Kermit, M.; Tomic, O. Independent Component Analysis Applied on Gas Sensor Array Measurement Data. IEEE Sens. J. 2003, 3, 218–228. [Google Scholar] [CrossRef]
- Scholz, M.; Gatzek, S.; Sterling, A.; Fiehn, O.; Selbig, J. Metabolite Fingerprinting: Detecting Biological Features by Independent Component Analysis. Bioinformatics 2004, 20, 2447–2454. [Google Scholar] [CrossRef] [Green Version]
- Worley, B.; Powers, R. PCA as a Practical Indicator of OPLS-DA Model Reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef]
- Liu, G.; Lee, D.P.; Schmidt, E.; Prasad, G.L. Pathway Analysis of Global Metabolomic Profiles Identified Enrichment of Caffeine, Energy, and Arginine Metabolism in Smokers but Not Moist Snuff. Bioinform. Biol. Insights 2019, 13, 1–11. [Google Scholar] [CrossRef]
- Richards, L.A. Diagnosis and Improvement of Saline and Alkaline Soils. Soil Sci. Soc. Am. J. 1954, 18, 348. [Google Scholar] [CrossRef]
- Qadir, M.; Schubert, S.; Steffens, D. Phytotoxic Substances in Soils. In Encyclopedia of Soils in the Environment; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar] [CrossRef]
- Paul, B.K.; Rashid, H.; Paul, B.K.; Rashid, H. Nonstructural Adaptation. In Climimatic Hazards in Coastal Bangladesh; Butterworth-Heinemann: Oxford, UK, 2017; pp. 209–256. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Im-Erb, R.; Neawsuparb, K.; Sombatpanit, S. Soil Salinization Assessment and Monitoring at Boe Klue District, Nan Province, Northern Thailand. In Developments in Soil Salinity Assessment and Reclamation: Innovative Thinking and Use of Marginal Soil and Water Resources in Irrigated Agriculture; Springer: Dordrecht, The Netherlands, 2013; pp. 75–86. [Google Scholar] [CrossRef]
- Arunin, S.; Pongwichian, P. Salt-Affected Soils and Management in Thailand. Bull. Soc. Sea Water Sci. Japan 2015, 69, 319–325. [Google Scholar] [CrossRef]
- Ambede, J.G.; Netondo, G.W.; Mwai, G.N.; Musyimi, D.M. NaCl Salinity Affects Germination, Growth, Physiology, and Biochemistry of Bambara Groundnut. Brazilian J. Plant Physiol. 2012, 24, 151–160. [Google Scholar] [CrossRef]
- Abreu, I.A.; Farinha, A.P.; Negrão, S.; Gonçalves, N.; Fonseca, C.; Rodrigues, M.; Batista, R.; Saibo, N.J.M.; Oliveira, M.M. Coping with Abiotic Stress: Proteome Changes for Crop Improvement. J. Proteom. 2013, 93, 145–168. [Google Scholar] [CrossRef]
- Sohrabi, S.; Ebadi, A.; Jalali, S.; Salami, S.A. Enhanced Values of Various Physiological Traits and VvNAC1 Gene Expression Showing Better Salinity Stress Tolerance in Some Grapevine Cultivars as Well as Rootstocks. Sci. Hortic. 2017, 225, 317–326. [Google Scholar] [CrossRef]
- Gil, R.; Boscaiu, M.; Lull, C.; Bautista, I.; Lidón, A.; Vicente, O. Are Soluble Carbohydrates Ecologically Relevant for Salt Tolerance in Halophytes? Funct. Plant Biol. 2013, 40, 805–818. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive Oxigen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-Induced Electrolyte Leakage: The Role of K+-Permeable Channels and Involvement in Programmed Cell Death and Metabolic Adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Bailey-Serres, J. The Roles of Reactive Oxygen Species in Plant Cells. Plant Physiol. 2006, 141, 311. [Google Scholar] [CrossRef]
- Caverzan, A.; Casassola, A.; Patussi Brammer, S. Reactive Oxygen Species and Antioxidant Enzymes Involved in Plant Tolerance to Stress. In Abiotic and Biotic Stress in Plants—Recent Advances and Future Perspectives; InTech: Rijeka, Croatia, 2016. [Google Scholar] [CrossRef]
- Ahmad, P.; Ashraf, M.; Hakeem, K.R.; Azooz, M.; Rasool, S.; Chandna, R.; Akram, N.A. Potassium Starvation-Induced Oxidative Stress and Antioxidant Defense Responses in Brassica Juncea. J. Plant Interact. 2014, 9, 1–9. [Google Scholar] [CrossRef]
- Mika, A.; Lüthje, S. Properties of Guaiacol Peroxidase Activities Isolated from Corn Root Plasma Membranes. Plant Physiol. 2003, 132, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Longet, D.; Penel, C.; Dunand, C. The Class III Peroxidase Multigenic Family in Rice and Its Evolution in Land. Plants Phytochemistry 2004, 65, 1879–1893. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New Insights on Plant Salt Tolerance Mechanisms and Their Potential Use for Breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Menezes-Benavente, L.; Teixeira, F.K.; Alvim Kamei, C.L.; Margis-Pinheiro, M. Salt Stress Induces Altered Expression of Genes Encoding Antioxidant Enzymes in Seedlings of a Brazilian Indica Rice (Oryza sativa L.). Plant Sci. 2004, 166, 323–331. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxidative Stress: Molecular Perception and Transduction of Signals Triggering Antioxidant Gene Defenses. Brazilian J. Med. Biol. Res. = Rev. Bras. Pesqui. medicas e Biol. 2005, 38, 995–1014. [Google Scholar] [CrossRef]
- Zaefizadeh, M.; Mohammad, S.; Jalali-E-Emam, S.; Alizadeh, B.; Zakarya, R.A.; Khayatnezhad, M. Superoxide Dismutase (SOD) Activity in Nacl Stress in Salt-Sensitive and Salt-Tolerance Genotypes of Colza (Brassica napus L.). Middle-East J. Sci. Res. 2011, 7, 7–11. [Google Scholar]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant Cellular and Molecular and Molecular Responses to High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Mittova, V.; Tal, M.; Volokita, M.; Guy, M. Up-Regulation of the Leaf Mitochondrial and Peroxisomal Antioxidative Systems in Response to Salt-Induced Oxidative Stress in the Wild Salt-Tolerant Tomato Species Lycopersicon pennellii. Plant. Cell Environ. 2003, 26, 845–856. [Google Scholar] [CrossRef]
- Demiral, T.; Turkan, I. Comparative Lipid Peroxidation, Antioxidant Defense Systems and Proline Content in Roots of Two Rice Cultivars Differing in Salt Tolerance. Environ. Exp. Bot. 2005, 53, 247–257. [Google Scholar] [CrossRef]
- El-Shabrawi, H.; Kumar, B.; Kaul, T.; Reddy, M.K.; Singla-Pareek, S.L.; Sopory, S.K. Redox Homeostasis, Antioxidant Defense, and Methylglyoxal Detoxification as Markers for Salt Tolerance in Pokkali Rice. Protoplasma 2010, 245, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Xu, S.; Hu, Q.; Mao, W.; Gong, Y. Putrescine Plays a Positive Role in Salt-Tolerance Mechanisms by Reducing Oxidative Damage in Roots of Vegetable Soybean. J. Integr. Agric. 2014, 13, 349–357. [Google Scholar] [CrossRef]
- Jiménez-Bremont, J.; Ruiz, O.; Rodríguez-Kessler, M. Modulation of Spermidine and Spermine Levels in Maize Seedlings Subjected to Long-Term Salt Stress. Plant Physiol. Biochem. PPB 2007, 45, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Newton, R.J. Polyamines Reduce Salt-Induced Oxidative Damage by Increasing the Activities of Antioxidant Enzymes and Decreasing Lipid Peroxidation in Virginia Pine. Plant Growth Regul. 2005, 46, 31–43. [Google Scholar] [CrossRef]
- Turan, M.A.; Turkmen, N.; Nilgun, T. Effect of NaCl on Stomatal Resistance and Proline, Chlorophyll, Na, Cl and K Concentrations of Lentil Plants. J. Agron. 2007, 6, 378–381. [Google Scholar] [CrossRef]
- Taffouo, V.D.; Wamba, O.F.; Youmbi, E.; Nono, G.V.; Akoa, A. Growth, Yield, Water Status and Ionic Distribution Response of Three Bambara Groundnut (Vigna subterranea (L.) Verdc.) Landraces Grown under Saline Conditions. Int. J. Bot. 2009, 6, 53–58. [Google Scholar] [CrossRef]
- Aarti, P.D.; Tanaka, R.; Tanaka, A. Effects of Oxidative Stress on Chlorophyll Biosynthesis in Cucumber (Cucumis sativus) Cotyledons. Physiol. Plant. 2006, 128, 186–197. [Google Scholar] [CrossRef]
- Zhao, G.Q.; Ma, B.L.; Ren, C.Z. Growth, Gas Exchange, Chlorophyll Fluorescence, and Ion Content of Naked Oat in Response to Salinity. Crop Sci. 2007, 47, 123. [Google Scholar] [CrossRef]
- Ashraf, M.; Ali, Q. Relative Membrane Permeability and Activities of Some Antioxidant Enzymes as the Key Determinants of Salt Tolerance in Canola (Brassica napus L.). Environ. Exp. Bot. 2008, 63, 266–273. [Google Scholar] [CrossRef]
- Zhang, H.; Ye, Y.K.; Wang, S.H.; Luo, J.P.; Tang, J.; Ma, D.F. Hydrogen Sulfide Counteracts Chlorophyll Loss in Sweetpotato Seedling Leaves and Alleviates Oxidative Damage against Osmotic Stress. Plant Growth Regul. 2009, 58, 243–250. [Google Scholar] [CrossRef]
- Walid, Z.; Houneida, A.; Najoua, M.; Chayma, O.; Mokhtar, L.; Zeineb, O. Photosynthetic Behaviour of Arabidopsis thaliana (Pa-1 Accession) under Salt Stress. African J. Biotechnol. 2013, 12, 4594–4602. [Google Scholar] [CrossRef]
- Heidari, M. Effects of Salinity Stress on Growth, Chlorophyll Content and Osmotic Components of Two Basil (Ocimum basilicum L.) Genotypes. African J. Biotechnol. 2012, 11, 379–384. [Google Scholar] [CrossRef]
- Gong, D.H.; Wang, G.Z.; Si, W.T.; Zhou, Y.; Liu, Z.; Jia, J. Effects of Salt Stress on Photosynthetic Pigments and Activity of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase in Kalidium Foliatum. Russ. J. Plant Physiol. 2018, 65, 98–103. [Google Scholar] [CrossRef]
- Ito, H.; Ohtsuka, T.; Tanaka, A. Conversion of Chlorophyll b to Chlorophyll a via 7-Hydroxymethyl Chlorophyll. J. Biol. Chem. 1996, 271, 1475–1479. [Google Scholar] [CrossRef] [Green Version]
- Mane, A.V.; Karadge, B.A.; Samant, J.S. Salinity Induced Changes in Photosynthetic Pigments and Polyphenols of Cymbopogon nardus (L.) Rendle. J. Chem. Pharm. Res. 2010, 2, 338–347. [Google Scholar]
- Elhaak, M.A.; Migahid, M.M.; Wegmann, K. Response on Photosynthetic Pigments to Drought and Salt Stress in Some Desert Species. Feddes Repert. 2008, 103, 573–577. [Google Scholar] [CrossRef]
- Gomes, A.D.C.; Pestana, M.; Abreu, I.; Claudete, S.-C.; Rachel Ann, H.-D.; Marina, S. Salinity Effects on Photosynthetic Pigments, Proline, Biomass and Nitric Oxide in Salvinia auriculata Aubl. Orig. Artic. Acta Limnol. Bras. 2017, 29, 9. [Google Scholar] [CrossRef]
- Wanke, M.; Skorupinska-Tudek, K.; Swiezewska, E. Isoprenoid Biosynthesis via 1-Deoxy-D-Xylulose 5-Phosphate/2-C-Methyl-D-Erythritol 4-Phosphate (DOXP/MEP) Pathway. Acta Biochim. Pol. 2001, 48, 663–672. [Google Scholar] [CrossRef]
- Munné-Bosch, S. The Role of Alpha-Tocopherol in Plant Stress Tolerance. J. Plant Physiol. 2005, 162, 743–748. [Google Scholar] [CrossRef]
- Che-Othman, M.; Jacoby, R.; Millar, A.; Taylor, N. Wheat Mitochondrial Respiration Shifts from the Tricarboxylic Acid Cycle to the GABA Shunt under Salt Stress. New Phytol. 2020, 225, 1166–1180. [Google Scholar] [CrossRef] [PubMed]
- Zidan, M.A.; Elewa, M.A. Effect of Salinity on Germination, Seedling Growth and Some Metabolic Changes in Four Plant Species (Umbelliferae). Indian J. Plant Physiol. 1995, 38, 57–61. [Google Scholar]
- Ribas-Carbo, M.; Taylor, N.L.; Giles, L.; Busquets, S.; Finnegan, P.M.; Day, D.A.; Lambers, H.; Medrano, H.; Berry, J.A.; Flexas, J. Effects of Water Stress on Respiration in Soybean Leaves. Plant Physiol. 2005, 139, 466. [Google Scholar] [CrossRef]
- Shekoofa, A.; Bijanzadeh, E.; Emam, Y.; Pessarakli, M. Effect of Salt Stress on Respipration of Various Wheat Lines/ Cultivars at Early Growth Stages. J. Plant Nutr. 2012, 36, 243–250. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of Salt Stress on Ion Balance and Nitrogen Metabolism of Old and Young Leaves in Rice (Oryza Sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, F.A.; Golovina, E.A.; Buitink, J. Mechanisms of Plant Desiccation Tolerance. Trends Plant Sci. 2001, 6, 431–438. [Google Scholar] [CrossRef]
- Seki, M.; Kamei, A.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Molecular Responses to Drought, Salinity and Frost: Common and Different Paths for Plant Protection. Curr. Opin. Biotechnol. 2003, 14, 194–199. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar Sensing and Signaling in Plants: Conserved and Novel Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Kempa, S.; Krasensky, J.; Dal Santo, S.; Kopka, J.; Jonak, C. A Central Role of Abscisic Acid in Stress-Regulated Carbohydrate Metabolism. PLoS ONE 2008, 3, e3935. [Google Scholar] [CrossRef]
- Farrant, J.M.; Moore, J.P. Programming Desiccation-Tolerance: From Plants to Seeds to Resurrection Plants. Curr. Opin. Plant Biol. 2011, 14, 340–345. [Google Scholar] [CrossRef]
- Gilbert, G.A.; Wilson, C.; Madore, M.A. Root-Zone Salinity Alters Raffinose Oligosaccharide Metabolism and Transport in Coleus. Plant Physiol. 1997, 115, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Dissecting the Roles of Osmolyte Accumulation during Stress. Plant Cell Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- Dong, S.; Beckles, D.M. Dynamic Changes in the Starch-Sugar Interconversion within Plant Source and Sink Tissues Promote a Better Abiotic Stress Response. J. Plant Physiol. 2019, 234–235, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Santelia, D. Starch as a Determinant of Plant Fitness under Abiotic Stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Dong, H.; Li, W.; Sun, Y.; Chen, S.; Kong, X. Increased Glycine Betaine Synthesis and Salinity Tolerance in AhCMO Transgenic Cotton Lines. Mol. Breed. 2009, 23, 289–298. [Google Scholar] [CrossRef]
- Bagdi, D.L.; Shaw, B.P. Analysis of Proline Metabolic Enzymes in Oryza Sativa under NaCl Stress. J. Environ. Biol. 2013, 34, 677–681. [Google Scholar]
- Huang, Z.; Zhao, L.; Chen, D.; Liang, M.; Liu, Z.; Shao, H.; Long, X. Salt Stress Encourages Proline Accumulation by Regulating Proline Biosynthesis and Degradation in Jerusalem Artichoke Plantlets. PLoS ONE 2013, 8, e62085. [Google Scholar] [CrossRef]
- Kausar, F.; Shahbaz, M. Interactive Effect of Foliar Application of Nitric Oxide (NO) and Salinity on Wheat (Triticum Aestivum L.). Pak. J. Bot 2013, 45, 67–73. [Google Scholar]
- Qu, X.; Wang, H.; Chen, M.; Liao, J.; Yuan, J.; Niu, G. Drought Stress–Induced Physiological and Metabolic Changes in Leaves of Two Oil Tea Cultivars. J. Am. Soc. Hortic. Sci. 2019, 144, 439–447. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Liu, C.; Li, S.; Zhu, D.; Zhao, Q.; Yu, J. Improved Nutritive Quality and Salt Resistance in Transgenic Maize by Simultaneously Overexpression of a Natural Lysine-Rich Protein Gene, SBgLR, and an ERF Transcription Factor Gene, TSRF1. Int. J. Mol. Sci. 2013, 14, 9459–9474. [Google Scholar] [CrossRef]
- Jespersen, D.; Yu, J.; Huang, B. Metabolic Effects of Acibenzolar-s-Methyl for Improving Heat or Drought Stress in Creeping Bentgrass. Front. Plant Sci. 2017, 8, 1224. [Google Scholar] [CrossRef] [PubMed]
- Kusvuran, S.; Dasgan, H.Y.; Abak, K. Citrulline Is an Important Biochemical Indicator in Tolerance to Saline and Drought Stresses in Melon. Sci. World J. 2013, 2013, 253414. [Google Scholar] [CrossRef] [PubMed]
- Joshi, V.; Joung, J.-G.; Fei, Z.; Jander, G. Interdependence of Threonine, Methionine and Isoleucine Metabolism in Plants: Accumulation and Transcriptional Regulation under Abiotic Stress. Amino Acids 2010, 39, 933–947. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Ouyang, C.; Wang, S.; Xu, Y.; Tang, L.; Chen, F. Effects of Salt Stress on Growth, Antioxidant Enzyme and Phenylalanine Ammonia-Lyase Activities in Jatropha curcas L. Seedlings. Plant, Soil Environ. 2008, 54, 374–381. [Google Scholar] [CrossRef]
- Haglund, B.M. Proline and Valine—Cues Which Stimulate Grasshopper Herbivory during Drought Stress? Nature 1980, 288, 697–698. [Google Scholar] [CrossRef]
- Khodary, S.E.A. Effect of Salinity and Tryptophan on Growth and Some Metabolic Changes in Wheat and Sorghum Plants. Biol. Plant. 1992, 34, 439–443. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Krishnan, S.R.; Pandian, S.; Mareeswaran, N.; Aruni, W.; Pandian, S.K.; Ramesh, M. Global Analysis of Threonine Metabolism Genes Unravel Key Players in Rice to Improve the Abiotic Stress Tolerance. Sci. Rep. 2018, 8, 9270. [Google Scholar] [CrossRef]
- Wu, D.; Cai, S.; Chen, M.; Ye, L.; Chen, Z.; Zhang, H.; Dai, F.; Wu, F.; Zhang, G. Tissue Metabolic Responses to Salt Stress in Wild and Cultivated Barley. PLoS ONE 2013, 8, e55431. [Google Scholar] [CrossRef]
- Hussein, M.M.; Gaballah, M.S.; El-Faham, S.Y. Amino Acids in Grains of Barley Plants as Affected by Benzyl Adenine and Salinity from Diluted Sea Water. J. Appl. Sci. 2005, 5, 655–658. [Google Scholar] [CrossRef] [Green Version]
- Sacchi, R.; Li, J.; Villarreal, F.; Gardell, A.M.; Kültz, D. Salinity-Induced Regulation of the Myo-Inositol Biosynthesis Pathway in Tilapia Gill Epithelium. J. Exp. Biol. 2013, 216, 4626–4638. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological Implications of Arginine Metabolism in Plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Zarei, A.; Brikis, C.J.; Bajwa, V.S.; Chiu, G.Z.; Simpson, J.P.; DeEll, J.R.; Bozzo, G.G.; Shelp, B.J. Plant Glyoxylate/Succinic Semialdehyde Reductases: Comparative Biochemical Properties, Function during Chilling Stress, and Subcellular Localization. Front. Plant Sci. 2017, 8, 1399. [Google Scholar] [CrossRef] [PubMed]
- Page, A.F.; Minocha, R.; Minocha, S.C. Living with High Putrescine: Expression of Ornithine and Arginine Biosynthetic Pathway Genes in High and Low Putrescine Producing Poplar Cells. Amino Acids 2012, 42, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Peng, F.; Xiao, Y.; Gong, Q.; Bao, Z.; Li, Y.; Wu, X. Mechanisms of High Concentration Valine-Mediated Inhibition of Peach Tree Shoot Growth. Front. Plant Sci. 2020, 11, 3067. [Google Scholar] [CrossRef] [PubMed]
- Bonner, C.A.; Rodrigues, A.M.; Miller, J.A.; Jensen, R.A. Amino Acids Are General Growth Inhibitors of Nicotiana Silvestris in Tissue Culture. Physiol. Plant. 1992, 84, 319–328. [Google Scholar] [CrossRef]
- Forsum, O.; Svennerstam, H.; Ganeteg, U.; Nsholm, T. Capacities and Constraints of Amino Acid Utilization in Arabidopsis. New Phytol. 2008, 179, 1058–1069. [Google Scholar] [CrossRef]
- Lea, P.J.; Sodek, L.; Parry, M.A.J.; Shewry, P.R.; Halford, N.G. Asparagine in Plants. Ann. Appl. Biol. 2007, 150, 1–26. [Google Scholar] [CrossRef]
- Hamberger, B.; Hahlbrock, K. The 4-Coumarate:CoA Ligase Gene Family in Arabidopsis Thaliana Comprises One Rare, Sinapate-Activating and Three Commonly Occurring Isoenzymes. Proc. Natl. Acad. Sci. USA 2004, 101, 2209–2214. [Google Scholar] [CrossRef]
- Maeda, H.; Yoo, H.; Dudareva, N. Prephenate Aminotransferase Directs Plant Phenylalanine Biosynthesis via Arogenate. Nat. Chem. Biol. 2011, 7, 19–21. [Google Scholar] [CrossRef]
- Allan, W.L.; Clark, S.M.; Hoover, G.J.; Shelp, B.J. Role of Plant Glyoxylate Reductases during Stress: A Hypothesis. Biochem. J. 2009, 423, 15–22. [Google Scholar] [CrossRef]
- Kiani-Pouya, A.; Roessner, U.; Jayasinghe, N.S.; Lutz, A.; Rupasinghe, T.; Bazihizina, N.; Bohm, J.; Alharbi, S.; Hedrich, R.; Shabala, S. Epidermal Bladder Cells Confer Salinity Stress Tolerance in the Halophyte Quinoa and Atriplex Species. Plant. Cell Environ. 2017, 40, 1900–1915. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Parida, A.K. Metabolomics and Network Analysis Reveal the Potential Metabolites and Biological Pathways Involved in Salinity Tolerance of the Halophyte Salvadora persica. Environ. Exp. Bot. 2018, 148, 85–99. [Google Scholar] [CrossRef]
- Liu, X.; Wu, H.; Ji, C.; Wei, L.; Zhao, J.; Yu, J. An Integrated Proteomic and Metabolomic Study on the Chronic Effects of Mercury in Suaeda salsa under an Environmentally Relevant Salinity. PLoS ONE 2013, 8, e64041. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhang, J.; Beckles, D.M. A Pivotal Role for Starch in the Reconfiguration of 14C-Partitioning and Allocation in Arabidopsis thaliana under Short-Term Abiotic Stress. Sci. Rep. 2018, 8, 9314. [Google Scholar] [CrossRef]
- Variyar, P.S.; Banerjee, A.; Akkarakaran, J.J.; Suprasanna, P. Role of Glucosinolates in Plant Stress Tolerance. In Emerging Technologies and Management of Crop Stress Tolerance; Elsevier: Amsterdam, The Netherlands, 2014; pp. 271–291. [Google Scholar] [CrossRef]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and Proteomics Reveal Drought-Stress Responses of Leaf Tissues from Spring-Wheat. Sci. Rep. 2018, 8, 5710. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jumpa, T.; Beckles, D.M.; Songsri, P.; Pattanagul, K.; Pattanagul, W. Physiological and Metabolic Responses of Gac Leaf (Momordica cochinchinensis (Lour.) Spreng.) to Salinity Stress. Plants 2022, 11, 2447. https://doi.org/10.3390/plants11192447
Jumpa T, Beckles DM, Songsri P, Pattanagul K, Pattanagul W. Physiological and Metabolic Responses of Gac Leaf (Momordica cochinchinensis (Lour.) Spreng.) to Salinity Stress. Plants. 2022; 11(19):2447. https://doi.org/10.3390/plants11192447
Chicago/Turabian StyleJumpa, Thitiwan, Diane M. Beckles, Patcharin Songsri, Kunlaya Pattanagul, and Wattana Pattanagul. 2022. "Physiological and Metabolic Responses of Gac Leaf (Momordica cochinchinensis (Lour.) Spreng.) to Salinity Stress" Plants 11, no. 19: 2447. https://doi.org/10.3390/plants11192447








