Multiomic Approaches Reveal Hormonal Modulation and Nitrogen Uptake and Assimilation in the Initial Growth of Maize Inoculated with Herbaspirillum seropedicae
Abstract
:1. Introduction
2. Results
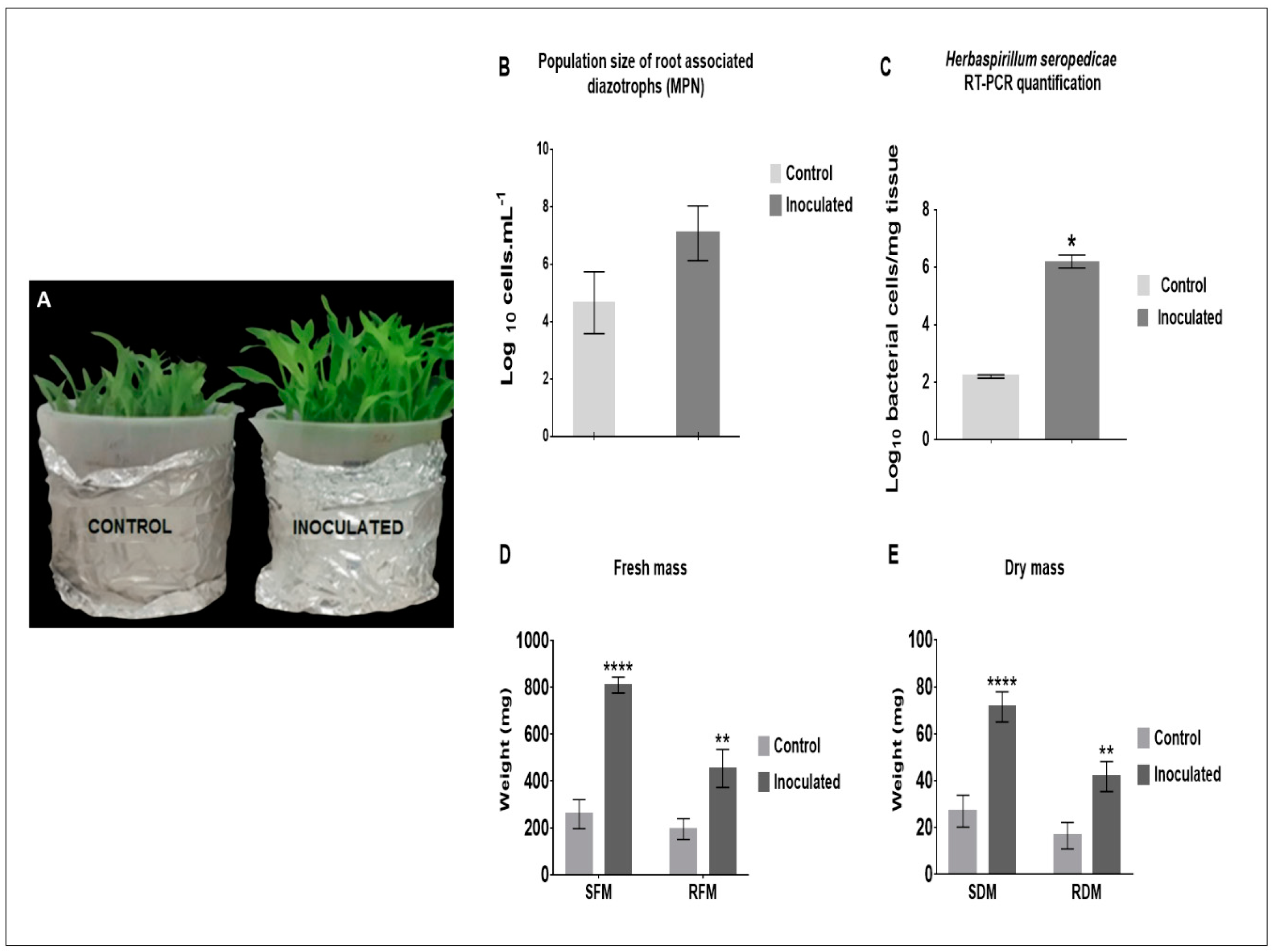
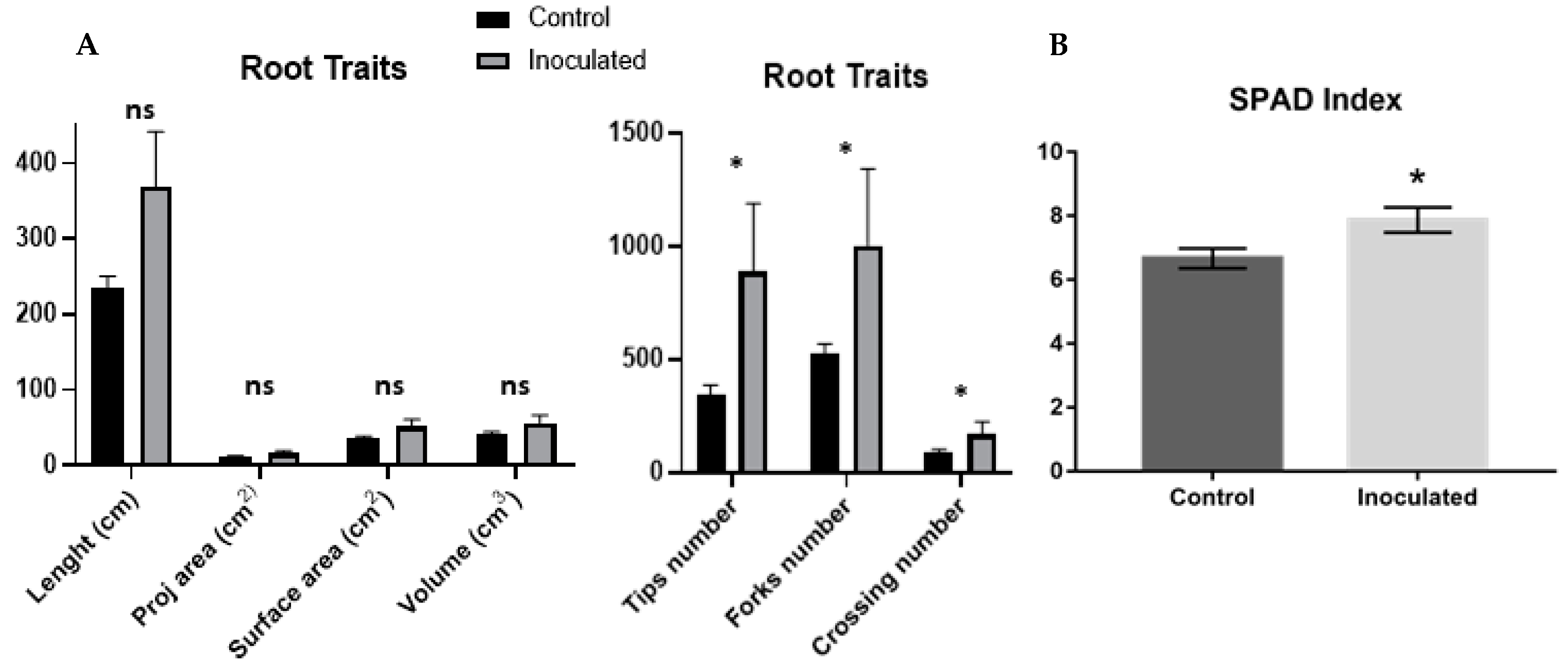
2.1. H. seropedicae Association and Plant Growth Promotion
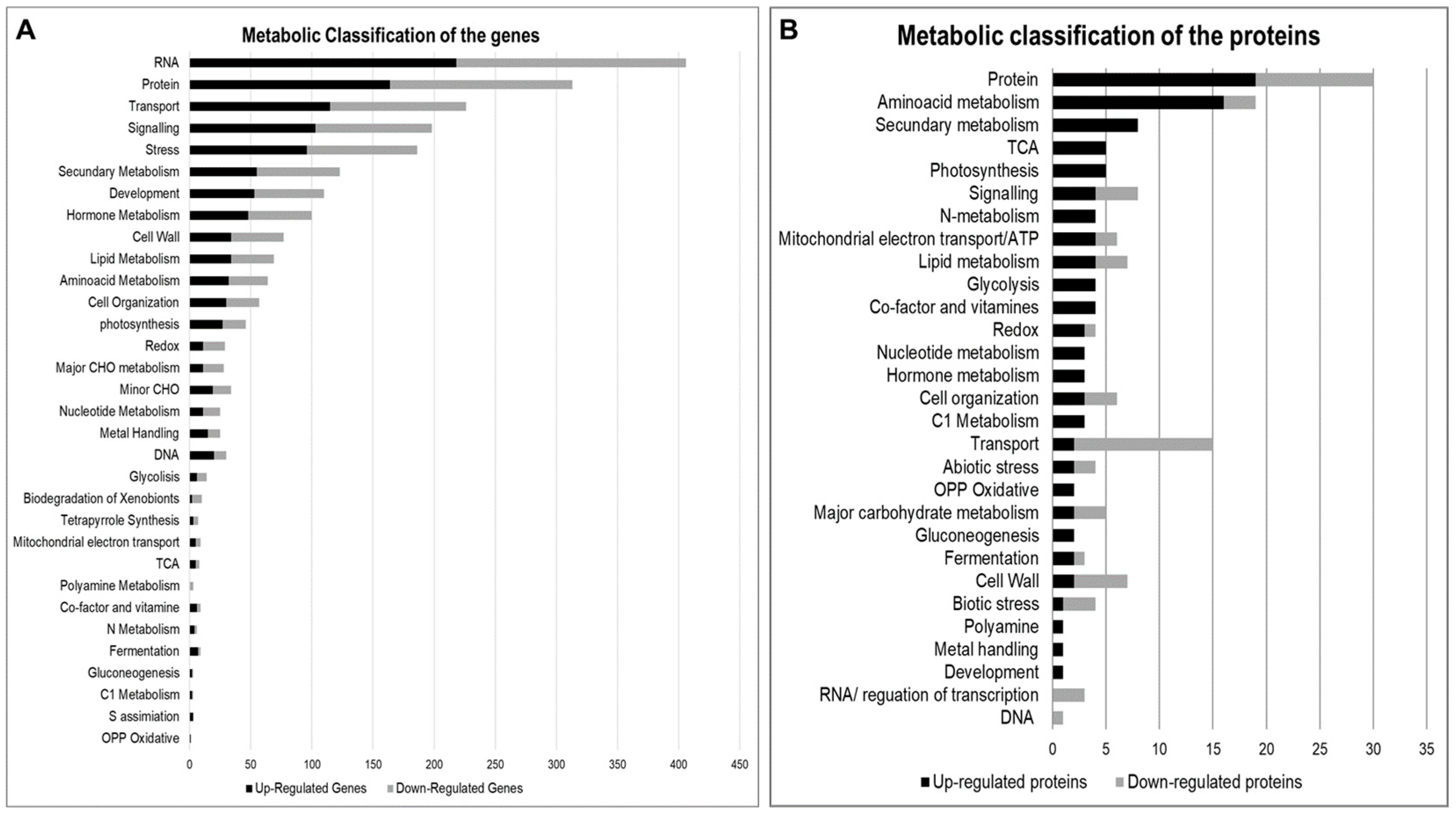
2.2. Multiomics Integrative Analysis
2.2.1. RNA Transcription
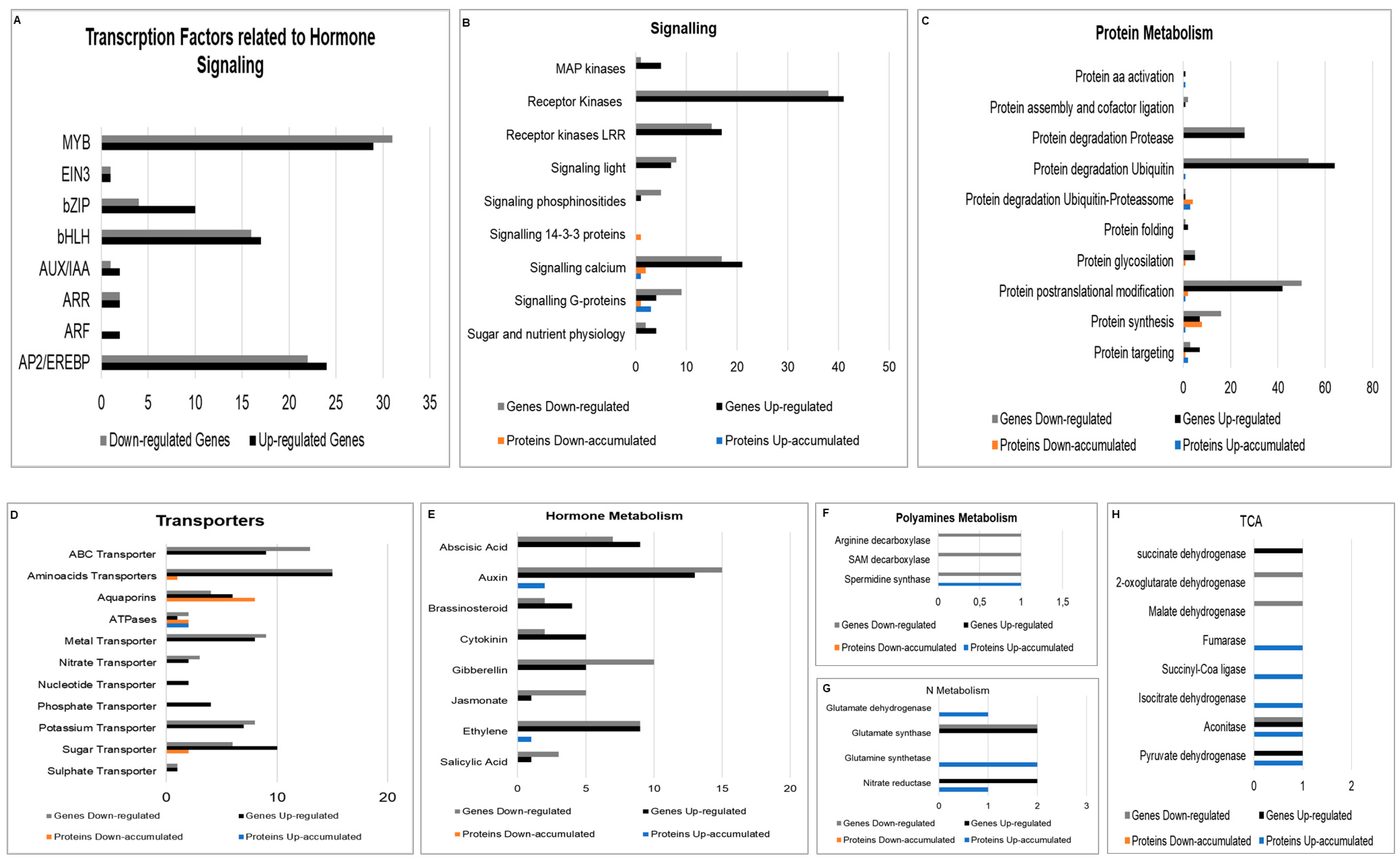
2.2.2. Signaling Related Genes
2.2.3. Protein Metabolism
2.2.4. Transporters
2.2.5. Hormone Regulation
2.2.6. Polyamine Metabolism
2.2.7. N and Amino Acid Metabolism
2.2.8. Energy Production
2.3. Validation of the DEGs and Proteins by Real-Time Quantitative PCR
2.4. Phytohormone Analyses
3. Discussion
3.1. Maize Growth Promotion and Hormonal Crosstalk
3.2. Modulation of the N and Amino Acids Metabolism
4. Materials and Methods
4.1. Preparation of Inoculum
4.2. Plant inoculation, Weight Measures and Root Morphology
4.3. Quantification of Diazotrophic Bacteria and H. seropedicae in Maize Roots
4.4. RNA Extraction and Sequencing
4.5. Bioinformatic Analysis of the Sequences Obtained by RNA-seq
4.6. Proteomic Procedures
4.6.1. Protein Extraction
4.6.2. Protein Digestion
4.6.3. Mass Spectrometry Analyses
4.6.4. Proteomics Data Analysis
4.7. Validation of DEGs and Proteins by Real-Time Quantitative PCR
4.8. Phytohormone Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pires, M.V.; da Cunha, D.A.; de Matos Carlos, S.; Costa, M.H. Nitrogen-use efficiency, nitrous oxide emissions, and cereal production in Brazil: Current Trends and Forecasts. PLoS ONE 2015, 10, e0135234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Withers, P.J.A.; Rodrigues, M.; Soltangheisi, A.; de Carvalho, T.S.; Guilherme, L.R.G.; Benites, V.M.; Gatiboni, L.C.; de Sousa, D.M.G.; Nunes, R.S.; Rosolem, C.A.; et al. Transitions to sustainable management of phosphorus in Brazilian agriculture. Sci. Rep. 2018, 8, 2537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [Green Version]
- Burney, J.A.; Davis, S.J.; Lobell, D.B. Greenhouse gas mitigation by agricultural intensification. Proc. Natl. Acad. Sci. USA 2010, 107, 12052–12057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toyota, K.; Watanabe, T. Recent trends in microbial inoculants in agriculture. Microbes. Environ. 2013, 28, 403–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldani, J.I.; Pot, B.; Kirchhof, G.; Falsen, E.; Baldani, V.L.; Olivares, F.L.; Hoste, B.; Kersters, K.; Hartmann, A.; Gillis, M.; et al. Emended description of Herbaspirillum; inclusion of [Pseudomonas] rubrisubalbicans, a milk plant pathogen, as Herbaspirillum rubrisubalbicans comb. nov.; and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. Int. J. Syst. Bacteriol. 1996, 46, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Olivares, F.L.; Baldani, V.L.D.; Reis, V.M.; Baldani, J.I.; Döbereiner, J. Occurrence of the endophytic diazotrophs Herbaspirillum spp. in roots, stems, and leaves, predominantly of Gramineae. Biol. Fertil. Soils 1996, 21, 197–200. [Google Scholar] [CrossRef]
- Baldotto, L.E.B.; Olivares, F.L.; Bressan-Smith, R. Structural interaction between GFP-labeled diazotrophic endophytic bacterium Herbaspirillum seropedicae RAM10 and pineapple plantlets’ Vitória’. Braz. J. Microbiol. 2011, 42, 114–125. [Google Scholar] [CrossRef]
- James, E.K.; Olivares, F.L. Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Crit. Rev. Plant Sci. 1998, 17, 77–119. [Google Scholar] [CrossRef]
- Monteiro, R.A.; Balsanelli, E.; Wassem, R.; Marin, A.M.; Brusamarello-Santos, L.C.C.; Schmidt, M.A.; Tadra-Sfeir, M.Z.; Pankievicz, V.; Cruz, L.M.; Chubatsu, L.S. Herbaspirillum-plant interactions: Microscopical, histological and molecular aspects. Plant Soil 2012, 356, 175–196. [Google Scholar] [CrossRef]
- James, E.K.; Gyaneshwar, P.; Mathan, N.; Barraquio, W.L.; Reddy, P.M.; Iannetta, P.P.; Olivares, F.L.; Ladha, J.K. Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant. Microbe. Interact. 2002, 15, 894–906. [Google Scholar] [CrossRef] [Green Version]
- Ramos, A.C.; Melo, J.; de Souza, S.B.; Bertolazi, A.A.; Silva, R.A.; Rodrigues, W.P.; Campostrini, E.; Olivares, F.L.; Eutropio, F.J.; Cruz, C.; et al. Inoculation with the endophytic bacterium Herbaspirillum seropedicae promotes growth, nutrient uptake and photosynthetic efficiency in rice. Planta 2020, 252, 87. [Google Scholar] [CrossRef]
- Roncato-Maccari, L.D.; Ramos, H.J.; Pedrosa, F.O.; Alquini, Y.; Chubatsu, L.S.; Yates, M.G.; Rigo, L.U.; Steffens, M.B.; Souza, E.M. Endophytic Herbaspirillum seropedicae expresses nif genes in gramineous plants. FEMS Microbiol. Ecol. 2003, 45, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, M.A.; Souza, E.M.; Baura, V.; Wassem, R.; Yates, M.G.; Pedrosa, F.O.; Monteiro, R.A. Evidence for the endophytic colonization of Phaseolus vulgaris (common bean) roots by the diazotroph Herbaspirillum seropedicae. Braz. J. Med. Biol. Res. 2011, 44, 182–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastián, F.; Cohen, A.; Piccoli, P.; Luna, V.; Baraldi, R.; Bottini, R. Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul. 1998, 24, 7–11. [Google Scholar] [CrossRef]
- Bottini, R.; Cassan, F.; Piccoli, P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 2004, 65, 497–503. [Google Scholar] [CrossRef]
- Radwan, T.E.-S.E.-D.; Mohamed, Z.K.; Reis, V.M. Efeito da inoculação de Azospirillum e Herbaspirillum na produção de compostos indólicos em plântulas de milho e arroz. Pesqui. Agropecuária Bras. 2004, 39, 987–994. [Google Scholar] [CrossRef]
- Estrada, G.A.; Baldani, V.L.D.; de Oliveira, D.M.; Urquiaga, S.; Baldani, J.I. Selection of phosphate-solubilizing diazotrophic Herbaspirillum and Burkholderia strains and their effect on rice crop yield and nutrient uptake. Plant Soil 2013, 369, 115–129. [Google Scholar] [CrossRef]
- Pedrosa, F.O.; Monteiro, R.A.; Wassem, R.; Cruz, L.M.; Ayub, R.A.; Colauto, N.B.; Fernandez, M.A.; Fungaro, M.H.; Grisard, E.C.; Hungria, M.; et al. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 2011, 7, e1002064. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.S.; Nogueira, M.A.; Hungria, M. Microbial inoculants: Reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. Amb Express 2019, 9, 1–22. [Google Scholar] [CrossRef]
- Klein, H.S.; Luna, F.V. The Impact of the Rise of Modern Maize Production in Brazil and Argentina. Hist. Agrar. Rev. Agric. História Rural 2022, 86, 273–310. [Google Scholar] [CrossRef]
- Alves, G.C.; Dos Santos, C.L.R.; Zilli, J.E.; Fabio, B.; Marriel, I.E.; Breda, F.A.d.F.; Boddey, R.M. Agronomic evaluation of Herbaspirillum seropedicae strain ZAE94 as an inoculant to improve maize yield in Brazil. Pedosphere 2021, 31, 583–595. [Google Scholar] [CrossRef]
- Araujo, E.d.O.; Mercante, F.M.; Vitorino, A.C.T.; Paim, L.R. Inoculation of Herbaspirillum seropedicae in three corn genotypes under different nitrogen levels. Embrapa Agropecuária Oeste-Artig. Periódico Indexado (ALICE) 2014. [Google Scholar]
- Canellas, L.P.; Balmori, D.M.; Médici, L.O.; Aguiar, N.O.; Campostrini, E.; Rosa, R.C.C.; Façanha, A.R.; Olivares, F.L. A combination of humic substances and Herbaspirillum seropedicae inoculation enhances the growth of maize (Zea mays L.). Plant Soil 2013, 366, 119–132. [Google Scholar] [CrossRef]
- Carvalho Dias, A.; Alves, G.C.; Da Silva, T.F.R.; Alves, B.J.R.; Santos, L.A.; Reis, V.M. Comparison of N uptake of maize inoculated with two diazotrophic bacterial species grown under two N levels. Arch. Agron. Soil Sci. 2022, 68, 1681–1697. [Google Scholar] [CrossRef]
- da Cunha, E.T.; Pedrolo, A.M.; Bueno, J.C.F.; Pereira, T.P.; Soares, C.; Arisi, A.C.M. Inoculation of Herbaspirillum seropedicae strain SmR1 increases biomass in maize roots DKB 390 variety in the early stages of plant development. Arch. Microbiol. 2022, 204, 373. [Google Scholar] [CrossRef]
- Dall’Asta, P.; Velho, A.C.; Pereira, T.P.; Stadnik, M.J.; Arisi, A.C.M. Herbaspirillum seropedicae promotes maize growth but fails to control the maize leaf anthracnose. Physiol. Mol. Biol. Plants 2019, 25, 167–176. [Google Scholar] [CrossRef]
- Pereira, T.P.; do Amaral, F.P.; Dall’Asta, P.; Brod, F.C.; Arisi, A.C. Real-time PCR quantification of the plant growth promoting bacteria Herbaspirillum seropedicae strain SmR1 in maize roots. Mol. Biotechnol. 2014, 56, 660–670. [Google Scholar] [CrossRef]
- Do Amaral, F.P.; Bueno, J.C.F.; Hermes, V.S.; Arisi, A.C.M. Gene expression analysis of maize seedlings (DKB240 variety) inoculated with plant growth promoting bacterium Herbaspirillum Seropedicae. Symbiosis 2014, 62, 41–50. [Google Scholar] [CrossRef]
- Hardoim, P.R.; de Carvalho, T.L.G.; Ballesteros, H.G.F.; Bellieny-Rabelo, D.; Rojas, C.A.; Venancio, T.M.; Ferreira, P.C.G.; Hemerly, A.S. Genome-wide transcriptome profiling provides insights into the responses of maize (Zea mays L.) to diazotrophic bacteria. Plant Soil 2020, 451, 121–143. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B.W. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 2000, 275, 3137–3143. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yue, R.; Tao, S.; Yang, Y.; Zhang, L.; Xu, M.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 2015, 57, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Rampey, R.A.; LeClere, S.; Kowalczyk, M.; Ljung, K.; Sandberg, G.r.; Bartel, B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004, 135, 978–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez Carranza, A.P.; Singh, A.; Steinberger, K.; Panigrahi, K.; Palme, K.; Dovzhenko, A.; Dal Bosco, C. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 2016, 6, 24212. [Google Scholar] [CrossRef] [Green Version]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef] [Green Version]
- Takato, S.; Kakei, Y.; Mitsui, M.; Ishida, Y.; Suzuki, M.; Yamazaki, C.; Hayashi, K.-i.; Ishii, T.; Nakamura, A.; Soeno, K. Auxin signaling through SCFTIR1/AFBs mediates feedback regulation of IAA biosynthesis. Biosci. Biotechnol. Biochem. 2017, 81, 1320–1326. [Google Scholar] [CrossRef] [Green Version]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold. Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef] [Green Version]
- Huttly, A.K.; Phillips, A.L. Gibberellin-regulated plant genes. Physiol. Plant 1995, 95, 310–317. [Google Scholar] [CrossRef]
- Sun, T.-p. Gibberellin signal transduction. Curr. Opin. Plant Biol. 2000, 3, 374–380. [Google Scholar] [CrossRef]
- Casimiro, I.; Marchant, A.; Bhalerao, R.P.; Beeckman, T.; Dhooge, S.; Swarup, R.; Graham, N.; Inze, D.; Sandberg, G.; Casero, P.J.; et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 2001, 13, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Olivares, F.L.; James, E.K.; Baldani, J.I.; Döbereiner, J. Infection of mottled stripe disease-susceptible and resistant sugar cane varieties by the endophytic diazotroph Herbaspirillum. New Phytol. 1997, 135, 723–737. [Google Scholar] [CrossRef]
- Knight, H.; Knight, M.R. Abiotic stress signalling pathways: Specificity and crosstalk. Trends Plant Sci. 2001, 6, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Poovaiah, B.W. Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 2003, 8, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, C.M.; Cura, J.A.; Cantore, M.L. Activation of a calcium-dependent protein kinase involved in the Azospirillum growth promotion in rice. World J. Microbiol. Biotechnol. 2017, 33, 22. [Google Scholar] [CrossRef]
- Ma, Q.-H. Small GTP-binding proteins and their functions in plants. J. Plant Growth Regul. 2007, 26, 369–388. [Google Scholar] [CrossRef]
- Palme, K.; Diefenthal, T.; Vingron, M.; Sander, C.; Schell, J. Molecular cloning and structural analysis of genes from Zea mays (L.) coding for members of the ras-related ypt gene family. Proc. Natl. Acad. Sci. USA 1992, 89, 787–791. [Google Scholar] [CrossRef] [Green Version]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 Proteins in Plant Hormone Signaling: Doing Several Things at Once. Front. Plant Sci. 2018, 9, 297. [Google Scholar] [CrossRef]
- Da Fonseca Breda, F.A.; da Silva, T.F.R.; Dos Santos, S.G.; Alves, G.C.; Reis, V.M. Modulation of nitrogen metabolism of maize plants inoculated with Azospirillum brasilense and Herbaspirillum seropedicae. Arch. Microbiol. 2019, 201, 547–558. [Google Scholar] [CrossRef]
- Kuang, W.; Sanow, S.; Kelm, J.M.; Muller Linow, M.; Andeer, P.; Kohlheyer, D.; Northen, T.; Vogel, J.P.; Watt, M.; Arsova, B. N-dependent dynamics of root growth and nitrate and ammonium uptake are altered by the bacterium Herbaspirillum seropedicae in the cereal model Brachypodium distachyon. J. Exp. Bot. 2022, 73, 5306–5321. [Google Scholar] [CrossRef]
- Nunes, R.O.; Domiciano Abrahao, G.; de Sousa Alves, W.; Aparecida de Oliveira, J.; Cesar Sousa Nogueira, F.; Pasqualoto Canellas, L.; Lopes Olivares, F.; Benedeta Zingali, R.; Soares, M.R. Quantitative proteomic analysis reveals altered enzyme expression profile in Zea mays roots during the early stages of colonization by Herbaspirillum seropedicae. Proteomics 2021, 21, e2000129. [Google Scholar] [CrossRef]
- Bredemeier, C.; Mundstock, C.M. Regulação da absorção e assimilação do nitrogênio nas plantas. Ciência Rural. 2000, 30, 365–372. [Google Scholar] [CrossRef]
- Curzi, M.J.; Ribaudo, C.M.; Trinchero, G.D.; Curá, J.A.; Pagano, E.A. Changes in the content of organic and amino acids and ethylene production of rice plants in response to the inoculation with Herbaspirillum seropedicae. J. Plant Interact. 2008, 3, 163–173. [Google Scholar] [CrossRef]
- Cañas, R.A.; Yesbergenova-Cuny, Z.; Belanger, L.; Rouster, J.; Brulé, L.; Gilard, F.; Quilleré, I.; Sallaud, C.; Hirel, B. NADH-GOGAT overexpression does not improve maize (Zea mays L.) performance even when pyramiding with NAD-IDH, GDH and GS. Plants 2020, 9, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oaks, A.; Hirel, B. Nitrogen assimilation in roots. Annu. Rev. Plant Physiol. 1985, 36, 345–365. [Google Scholar] [CrossRef]
- Fontaine, J.X.; Terce-Laforgue, T.; Armengaud, P.; Clement, G.; Renou, J.P.; Pelletier, S.; Catterou, M.; Azzopardi, M.; Gibon, Y.; Lea, P.J.; et al. Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 2012, 24, 4044–4065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelp, B.J.; Bozzo, G.G.; Trobacher, C.P.; Zarei, A.; Deyman, K.L.; Brikis, C.J. Hypothesis/review: Contribution of putrescine to 4-aminobutyrate (GABA) production in response to abiotic stress. Plant Sci. 2012, 193–194, 130–135. [Google Scholar] [CrossRef]
- Altman, A.; Levin, N. Interactions of polyamines and nitrogen nutrition in plants. Physiol. Plant 1993, 89, 653–658. [Google Scholar] [CrossRef]
- Majumdar, R.; Barchi, B.; Turlapati, S.A.; Gagne, M.; Minocha, R.; Long, S.; Minocha, S.C. Glutamate, Ornithine, Arginine, Proline, and Polyamine Metabolic Interactions: The Pathway Is Regulated at the Post-Transcriptional Level. Front. Plant. Sci. 2016, 7, 78. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.; Jiang, Y.; Liu, L.; Zhang, Z.; Zheng, Y. Identification of transcriptome induced in roots of maize seedlings at the late stage of waterlogging. BMC Plant Biol. 2010, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Canellas, N.O.A.; Olivares, F.L.; Canellas, L.P. Metabolite fingerprints of maize and sugarcane seedlings: Searching for markers after inoculation with plant growth-promoting bacteria in humic acids. Chem. Biol. Technol. Agric. 2019, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Booij, R.; Valenzuela, J.L.; Aguilera, C. Determination of crop nitrogen status using non-invasive methods. Management of Nitrogen and Water in Potato Production; Wageningen Pers: Wageningen, The Netherlands, 2000; pp. 72–82. [Google Scholar]
- Andersson, J.; Wentworth, M.; Walters, R.G.; Howard, C.A.; Ruban, A.V.; Horton, P.; Jansson, S. Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II—Effects on photosynthesis, grana stacking and fitness. Plant J. 2003, 35, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Dembinsky, D.; Woll, K.; Saleem, M.; Liu, Y.; Fu, Y.; Borsuk, L.A.; Lamkemeyer, T.; Fladerer, C.; Madlung, J.; Barbazuk, B.; et al. Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 2007, 145, 575–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.G.; Li, B.; Liu, S.X.; Wang, H.W.; Li, C.P.; Song, S.H.; Beatty, M.; Zastrow-Hayes, G.; Yang, X.H.; Qin, F.; et al. Characterization of Proteome Variation During Modern Maize Breeding. Mol. Cell. Proteomics 2019, 18, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Walley, J.W.; Sartor, R.C.; Shen, Z.; Schmitz, R.J.; Wu, K.J.; Urich, M.A.; Nery, J.R.; Smith, L.G.; Schnable, J.C.; Ecker, J.R.; et al. Integration of omic networks in a developmental atlas of maize. Science 2016, 353, 814–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldani, J.I.; Reis, V.M.; Videira, S.S.; Boddey, L.H.; Baldani, V.L.D. The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semisolid media: A practical guide for microbiologists. Plant Soil 2014, 384, 413–431. [Google Scholar] [CrossRef]
- Fernandes, B.S.P.M.; de Araújo, J.L.S.; Varial, L.H.; Baldanib, V.L.D. Development of a real-time PCR assay for the detection and quantification of Gluconacetobacter diazotrophicus in sugarcane grown under field conditions. Afr. J. Microbiol. Res. 2014, 8, 2937–2946. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2017. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 July 2019).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Thimm, O.; Blasing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Kruger, P.; Selbig, J.; Muller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Damerval, C.; De Vienne, D.; Zivy, M.; Thiellement, H. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 1986, 7, 52–54. [Google Scholar] [CrossRef]
- Nanjo, Y.; Skultety, L.; Uvackova, L.; Klubicova, K.; Hajduch, M.; Komatsu, S. Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J. Proteome. Res. 2012, 11, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Burrieza, H.P.; Rizzo, A.J.; Moura Vale, E.; Silveira, V.; Maldonado, S. Shotgun proteomic analysis of quinoa seeds reveals novel lysine-rich seed storage globulins. Food Chem. 2019, 293, 299–306. [Google Scholar] [CrossRef]
- Almeida, F.A.; Vale, E.M.; Reis, R.S.; Santa-Catarina, C.; Silveira, V. LED lamps enhance somatic embryo maturation in association with the differential accumulation of proteins in the Carica papaya L. ‘Golden’ embryogenic callus. Plant Physiol. Biochem. 2019, 143, 109–118. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 2014, 11, 167–170. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Oliveros, J.C.; VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 3 May 2020).
- Lin, Y.; Zhang, C.; Lan, H.; Gao, S.; Liu, H.; Liu, J.; Cao, M.; Pan, G.; Rong, T.; Zhang, S. Validation of potential reference genes for qPCR in maize across abiotic stresses, hormone treatments, and tissue types. PLoS ONE 2014, 9, e95445. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Filgueiras, L.; Silva, R.; Almeida, I.; Vidal, M.; Baldani, J.I.; Meneses, C.H.S.G. Gluconacetobacter diazotrophicus mitigates drought stress in Oryza sativa L. Plant Soil 2020, 451, 57–73. [Google Scholar] [CrossRef]
- Silva, R.; Filgueiras, L.; Santos, B.; Coelho, M.; Silva, M.; Estrada-Bonilla, G.; Vidal, M.; Baldani, J.I.; Meneses, C. Gluconacetobacter diazotrophicus Changes the Molecular Mechanisms of Root Development in Oryza sativa L. Growing Under Water Stress. Int. J. Mol. Sci. 2020, 21, 333. [Google Scholar] [CrossRef] [PubMed]


| Samples | Uniquely Mapped | Mapped to Multiple Loci | Mapped to too Many Loci | Unmapped: Too Short | Unmapped: Other | % of Mapped Genes |
|---|---|---|---|---|---|---|
| Control 1 | 76,454,565 | 4,169,270 | 462,100 | 6,955,351 | 35,217 | 86.80 |
| Control 2 | 27,685,993 | 1,565,491 | 55,449 | 2,497,993 | 19,093 | 86.99 |
| Control 3 | 51,901,676 | 2,850,385 | 260,875 | 4,381,316 | 29,724 | 87.34 |
| Inoculated 1 | 34,434,572 | 2,451,679 | 754,582 | 14,461,553 | 36,493 | 66.04 |
| Inoculated 2 | 13,506,360 | 806,095 | 27,396 | 1,936,178 | 13,038 | 82.91 |
| Inoculated 3 | 33,860,470 | 2,281,374 | 324,017 | 10,106,497 | 32,632 | 72.65 |
| RNA-Seq | Proteomic | RT-qPCR | |
|---|---|---|---|
| b6f | 1.88 | - | 5.41 |
| CALM1 | - | 1.62 | 4.55 |
| GID1L2 | - | 0.6 | 4.08 |
| PsbO_1 | 0.14 | - | 3.79 |
| GAST1 | 0.03 | - | 3.39 |
| PHTP9 | 0.15 | - | 2.40 |
| CAB-1 | 2.12 | - | 2.25 |
| PIP1-5 | 1.01 | −1.1 | 2.02 |
| AO2 | - | 0.84 | −2.34 |
| CHOR | - | 0.68 | 0.17 |
| HMTD | −0.63 | - | −0.40 |
| Root | Shoots | |||
|---|---|---|---|---|
| Uninoculated | Inoculated | Uninoculated | Inoculated | |
| IAA | 15.49 ± 0.41 B | 29.63 ± 0.51 A | 15.34 ± 0.39 B | 19.57 ± 0.37 A |
| IBA | 11.13 ± 0.72 B | 14.62 ± 0.24 A | 11.37 ± 0.43 B | 12.08 ± 0.17 B |
| 4-Cl-IAA | 4.3 ± 0.35 B | 8.67 ± 0.33 A | 4.35 ± 0.35 B | 4.67 ± 0,33 B |
| GA1 | 8.73 ± 0.35 B | 11.91 ± 0.19 A | 9.045 ± 0.05 B | 16.49 ± 0.29 A |
| GA3 | 14.84 ± 0.16 B | 20.46 ± 0.29 A | 14.67 ± 0.37 B | 29.02 ± 1.14 A |
| T-ZEATIN | 8.46 ± 0.41 B | 10.56 ± 0.26 A | 8.55 ± 0.25 B | 14.27 ± 0.38 A |
| ABA | 18.42 ± 0.44 B | 21.41 ± 0.40 A | 19.42 ± 0.21 B | 25.35 ± 0.27 A |
| Brassinosteroids | 3.22 ± 0.14 B | 5.26 ± 0.50 A | 4.42 ± 0.25 B | 7.72 ± 0.24 A |
| Salicylic acid | 1.70 ± 0.26 B | 5.39 ± 0.21 A | 2.68 ± 0.21 B | 2.59 ± 0.50 B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irineu, L.E.S.d.S.; Soares, C.d.P.; Soares, T.S.; Almeida, F.A.d.; Almeida-Silva, F.; Gazara, R.K.; Meneses, C.H.S.G.; Canellas, L.P.; Silveira, V.; Venancio, T.M.; et al. Multiomic Approaches Reveal Hormonal Modulation and Nitrogen Uptake and Assimilation in the Initial Growth of Maize Inoculated with Herbaspirillum seropedicae. Plants 2023, 12, 48. https://doi.org/10.3390/plants12010048
Irineu LESdS, Soares CdP, Soares TS, Almeida FAd, Almeida-Silva F, Gazara RK, Meneses CHSG, Canellas LP, Silveira V, Venancio TM, et al. Multiomic Approaches Reveal Hormonal Modulation and Nitrogen Uptake and Assimilation in the Initial Growth of Maize Inoculated with Herbaspirillum seropedicae. Plants. 2023; 12(1):48. https://doi.org/10.3390/plants12010048
Chicago/Turabian StyleIrineu, Luiz Eduardo Souza da Silva, Cleiton de Paula Soares, Tatiane Sanches Soares, Felipe Astolpho de Almeida, Fabrício Almeida-Silva, Rajesh Kumar Gazara, Carlos Henrique Salvino Gadelha Meneses, Luciano Pasqualoto Canellas, Vanildo Silveira, Thiago Motta Venancio, and et al. 2023. "Multiomic Approaches Reveal Hormonal Modulation and Nitrogen Uptake and Assimilation in the Initial Growth of Maize Inoculated with Herbaspirillum seropedicae" Plants 12, no. 1: 48. https://doi.org/10.3390/plants12010048









