Establishment of a Rapid and Effective Agrobacterium-Mediated Genetic Transformation System of Oxalis triangularis ‘Purpurea’
Abstract
:1. Introduction
2. Results
2.1. Different Recipient Explants of O. triangularis ‘Purpurea’ Showed Different Transformation Efficiency
2.2. The Effects of Antibiotic Concentrations on Petiole Growth
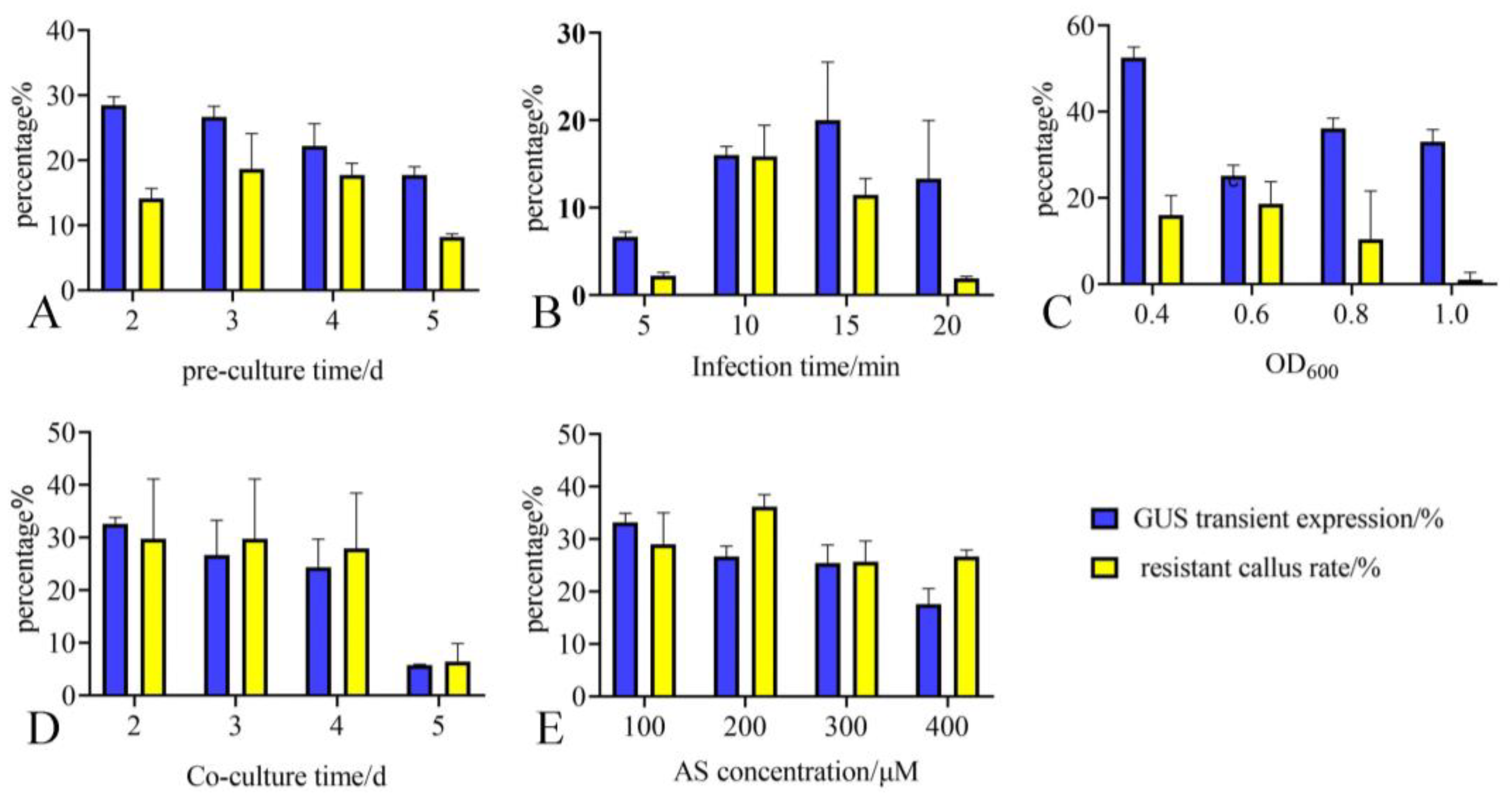
2.3. Optimizing Key Parameters by Combining Transient Expression Rate and Resistant Callus Rate
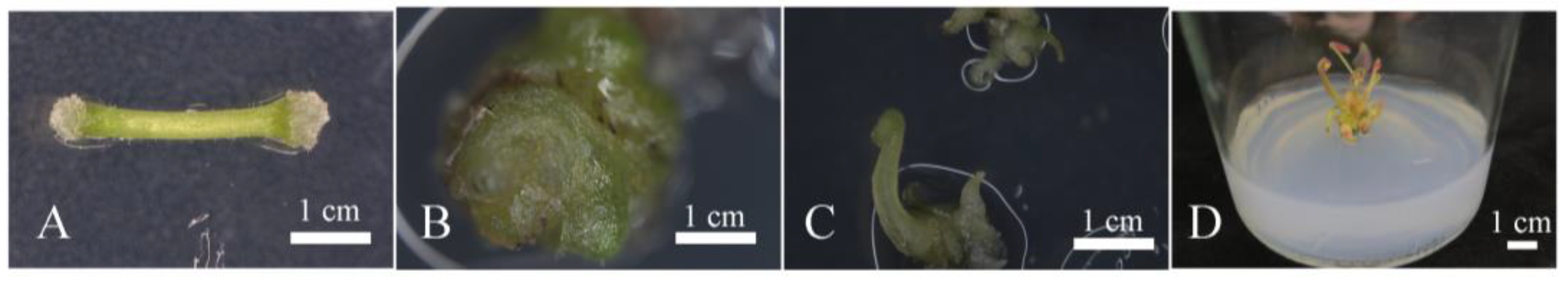
2.4. Regeneration of Hygromycin-Resistant Plants
2.5. Determination of Genetic Transformation Efficiency
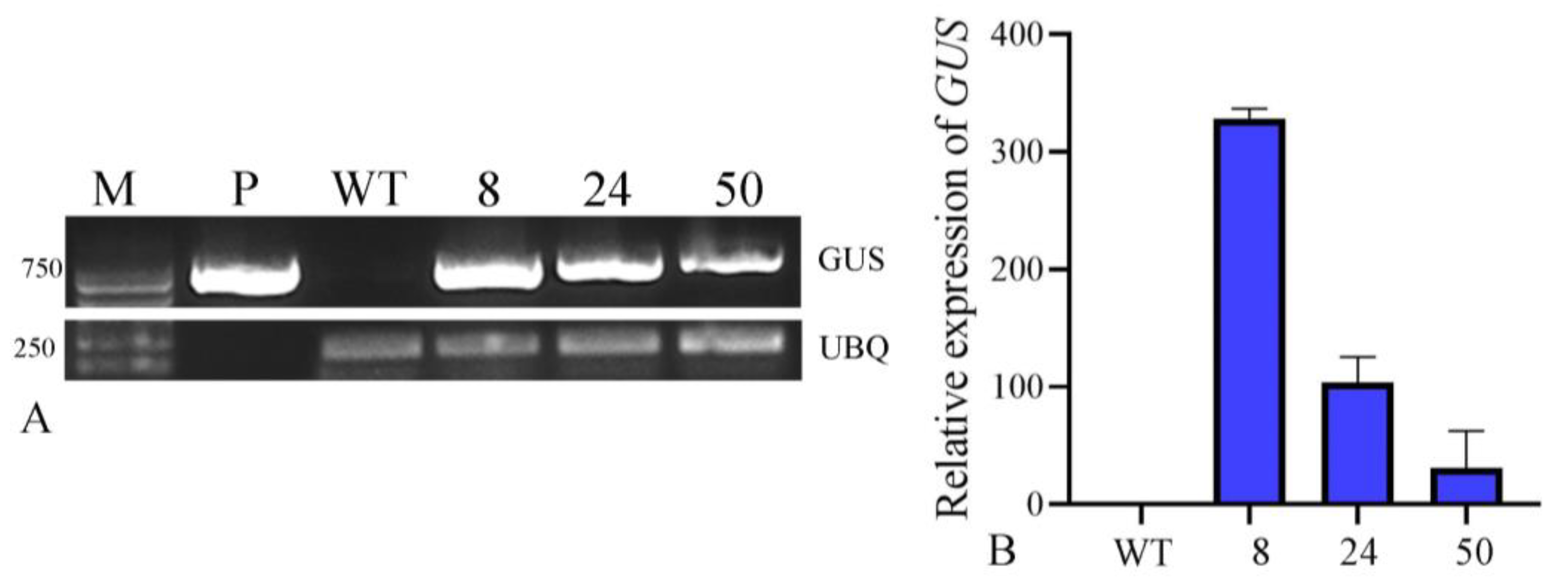
2.5.1. GUS Staining of the Hygromycin-Resistant Plants of O. triangularis ‘Purpurea’
2.5.2. PCR Identification of the Hygromycin-Resistant Plants of O. triangularis ‘Purpurea’
2.5.3. Detection of Transgene Expression by RT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Medium
4.2. Plant Expression Vector and Agrobacterium Strain
4.3. Determination of Transformation Receptor
4.4. Determination of Selection Pressure
4.5. Optimization of the Parameters for Genetic Transformation
4.6. The GUS Staining
4.7. PCR Identification of Positive Plants
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosna, M.T.; Noraini, M.; Jamilah, S.Y.; Noorlidah, A.; Sadegh, M. Synthetic seeds production and regeneration of Oxalis triangularis for mass propagation and conservation. J. Int. Environ. Appl. Sci. 2013, 4, 461–464. [Google Scholar]
- Rojas, C.; Díaz, G.; Montenegro, O.; Ferrada, E. Occurrence of leaf rust disease in Oxalis triangularis caused by Puccinia oxalidis in Valdivia, Chile. Plant Dis. 2023, 107, 2225. [Google Scholar] [CrossRef]
- Luo, B.; Chen, L.; Chen, G.; Wang, Y.; Xie, Q.; Chen, X.; Hu, Z. Transcription and metabolism pathways of anthocyanin in purple shamrock (Oxalis triangularis A. St.-Hil.). Metabolites 2022, 12, 1290. [Google Scholar] [CrossRef]
- Xia, P.; Hu, W.; Liang, T.; Yang, D.; Liang, Z. An attempt to establish an Agrobacterium-mediated transient expression system in medicinal plants. Protoplasma 2020, 257, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.; Han, J.E.; Ho, T.-T.; Park, S.Y. Development of hairy root cultures for biomass and triterpenoid production in Centella asiatica. Plants 2022, 11, 148. [Google Scholar] [CrossRef]
- Su, W.B.; Xu, M.Y.; Radani, Y.; Yang, L.M. Technological development and application of plant genetic transformation. Int. J. Mol. Sci. 2023, 24, 10646. [Google Scholar] [CrossRef]
- Gupta, V.; Rahman, L.-U. An efficient plant regeneration and Agrobacterium-mediated genetic transformation of Tagetes erecta. Protoplasma 2015, 252, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, D.; Long, X.; Hao, Z.; Lu, Y.; Zhou, Y.; Peng, Y.; Cheng, T.; Shi, J.; Chen, J. Agrobacterium-mediated genetic transformation of embryogenic callus in a Liriodendron hybrid (L. Chinense × L. Tulipifera). Front. Plant Sci. 2022, 13, 705. [Google Scholar] [CrossRef]
- Jin, C.L.; Dong, L.Q.; Wei, C.; Wani, M.A.; Yang, C.M.; Li, S.C.; Li, F. Creating novel ornamentals via new strategies in the era of genome editing. Front. Plant Sci. 2023, 14, 1142866. [Google Scholar] [CrossRef]
- Chen, Y.; Hou, X.; Zheng, Y.; Lyu, Y. The establishment of a genetic transformation system and the acquisition of transgenic plants of oriental hybrid lily (Lilium L.). Int. J. Mol. Sci. 2023, 24, 782. [Google Scholar] [CrossRef]
- Zvi, M.M.B.; Negre-Zakharov, F.; Masci, T.; Ovadis, M.; Shklarman, E.; Ben-Meir, H.; Tzfira, T.; Dudareva, N.; Vainstein, A. Interlinking showy traits: Co-engineering of scent and colour biosynthesis in flowers. Plant Biotechnol. J. 2008, 6, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Zvi, M.M.B.; Shklarman, E.; Masci, T.; Kalev, H.; Debener, T.; Shafir, S.; Ovadis, M.; Vainstein, A. PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytol. 2012, 195, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Chilton, M.-D.; Tepfer, D.-A.; Petit, A.; David, C.; Casse-Delbart, F.; Tempé, J. Agrobacterium rhizogenes inserts T-DNA into the genomes of the host plant root cells. Nature 1982, 295, 432–434. [Google Scholar] [CrossRef]
- Canto, T. Transient expression systems in plants: Potentialities and constraints. Adv. Exp. Med. Biol. 2016, 896, 287–301. [Google Scholar]
- Idnurm, A.; Bailey, A.M.; Cairns, T.C.; Elliott, C.E.; Foster, G.D.; Ianiri, G.; Jeon, J. A silver bullet in a golden age of functional genomics: The impact of Agrobacterium-mediated transformation of fungi. Fungal Biol. Biotech. 2017, 4, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Mudunkothge, J.S.; Krizek, B.A. The GUS reporter system in flower development studies. In Flower Development. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2014; pp. 295–304. [Google Scholar]
- De Ruijter, N.C.A.; Verhees, J.; van Leeuwen, W.; van der Krol, A.R. Evaluation and comparison of the GUS, LUC and GFP reporter system for gene expression studies in plants. Plant Biol. 2003, 5, 103–115. [Google Scholar] [CrossRef]
- Yan, R.; Wang, Z.P.; Ren, Y.M.; Li, H.Y.; Liu, N.; Sun, H.M. Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. Int. J. Mol. Sci. 2019, 20, 2920. [Google Scholar] [CrossRef]
- Qiu, J.; Sun, S.; Luo, S.; Zhang, J.; Xiao, X.; Zhang, L.; Wang, F.; Liu, S. Arabidopsis AtPAP1 transcription factor induces anthocyanin production in transgenic Taraxacum brevicorniculatum. Plant Cell Rep. 2014, 33, 669–680. [Google Scholar] [CrossRef]
- Xu, T.; Wu, D.; Yang, X.; Fu, H.; Wang, J.G. Optimization of genetic transformation system of Oxalis triangularis ‘Purpurea’. Heilongjiang Sci. 2010, 1, 1–3+11. [Google Scholar]
- Tie, W.; Zhou, F.; Wang, L.; Xie, W.; Chen, H.; Li, X.; Lin, Y. Reasons for lower transformation efficiency in indica rice using Agrobacterium tumefaciens-mediated transformation: Lessons from transformation assays and genome-wide expression profiling. Plant Mol. Biol. 2012, 78, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Alaguero-Cordovilla, A.; Sánchez-García, A.B.; Ibáñez, S.; Albacete, A.; Cano, A.; Acosta, M.; Pérez-Pérez, J.M. An auxin-mediated regulatory framework for wound-induced adventitious root formation in tomato shoot explants. Plant Cell Environ. 2021, 44, 1642–1662. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Sheng, X.; Yu, H.; Wang, J.; Shen, Y.; Gu, H. Establishment of a stable, effective and universal genetic transformation technique in the diverse species of Brassica oleracea. Front. Plant. Sci. 2022, 13, 1021669. [Google Scholar] [CrossRef] [PubMed]
- Supartana, P.; Shimizu, T.; Shioiri, H.; Nogawa, M.; Nozue, M.; Kojima, M. Development of simple and efficient in planta transformation method for rice (Oryza sativa L.) using Agrobacterium tumefaciens. J. Biosci. Bioeng. 2005, 100, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Chhikara, S.; Chaudhary, D.; Yadav, M.; Sainger, M.; Jaiwal, P.-K. A non-tissue culture approach for developing transgenic Brassica juncea L. plants with Agrobacterium tumefaciens. In Vitro Cell. Dev. Biol. Plant 2012, 48, 7–14. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, J.; Hao, X.; Xie, Y.; Lv, K.; Xu, W. Establishment of a stable grape immature zygotic embryo-based genetic transformation system. Sci. Hortic. 2023, 316, 112009. [Google Scholar] [CrossRef]
- Wilmink, A.; Dons, J. Selective agents and marker genes for use in transformation of monocotyledonous plants. Plant Mol. Biol. Rep. 1993, 11, 165–185. [Google Scholar] [CrossRef]
- Wang, X.H.; Zhou, F.L.; Liu, J.L.; Liu, W.Q.; Zhang, S.L.; Li, D.L.; Song, J.K.; Wang, R.; Yang, Y.J. Establishment of efficient callus genetic transformation system for Pyrus armeniacaefolia. Sci. Hortic. 2021, 289, 110429. [Google Scholar] [CrossRef]
- Chen, J.; Wang, L.; Chen, J.B.; Huang, J.; Liu, F.; Guo, R.; Yang, L.; Grabon, A.; Zhao, K.; Kong, F.L.; et al. Agrobacterium tumefaciens-mediated transformation system for the important medicinal plant Dendrobium catenatum Lindl. In Vitro Cell. Dev. Biol. Plant 2018, 54, 228–239. [Google Scholar] [CrossRef]
- Yao, J.L.; Tomes, S.; Gleave, A.P. Transformation of apple (Malus× domestica) using mutants of apple acetolactate synthase as a selectable marker and analysis of the T-DNA integration sites. Plant Cell Tissue Organ Cult. 2013, 32, 703–714. [Google Scholar] [CrossRef]
- Xiao, X.; Li, Y.; Qin, J.L.; He, Y.; Cai, W.Y.; Chen, Z.W.; Xi, L.Y.; Zhang, J.M. An optimized Agrobacterium tumefaciens-mediated transformation system for random insertional mutagenesis in Fonsecaea monophora. J. Microbiol. Methods. 2020, 170, 105838. [Google Scholar] [CrossRef]
- Singh, P.; Khan, S.; Kumar, S.; ur Rahman, L. Establishment of an efficient Agrobacterium-mediated genetic transformation system in Pelargonium graveolens: An important aromatic plant. Plant Cell Tissue Organ Cult. 2017, 129, 35–44. [Google Scholar] [CrossRef]
- Huang, P.; Xu, M.; Xia, L.; Qing, Z.; Tang, Q.; Liu, W.; Zeng, J. Establishment of an efficient Agrobacterium-mediated genetic transformation method in Macleaya cordata. Sci. Hortic. 2017, 226, 302–306. [Google Scholar] [CrossRef]
- Du, Z.Z.; Zong, Q.Q.; Gao, H.F.; Guo, Q.Y.; Liu, T.G.; Chen, W.Q.; Gao, L. Development of an Agrobacterium tumefaciens-mediated transformation system for Tilletia controversa Kuhn. J. Microbiol. Methods 2021, 189, 106313. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Ishida, Y.; Komari, T. Progress of cereal transformation technology mediated by Agrobacterium tumefaciens. Front. Plant Sci. 2014, 5, 628. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Ji, J.J.; Jiang, F.; Tian, X.R.; Li, J.K.; Gao, J.P. Establishment of a genetic transformation system for Codonopsis pilosula callus. Plant Biotechnol. 2022, 39, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.T.; Chang, H.H.; Ho, S.L.; Tong, W.F.; Yu, S.M. Agrobacterium-mediated production of transgenic rice plants expressing a chimeric α-amylase promoter/β-glucuronidase gene. Plant Mol. Biol. 1993, 22, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Ogaki, M.; Furuichi, Y.; Kuroda, K.; Chin, D.; Ogawa, Y.; Mii, M. Importance of co-cultivation medium pH for successful Agrobacterium-mediated transformation of Lilium× formolongi. Plant Cell Rep. 2008, 27, 699–705. [Google Scholar] [CrossRef]
- Nakano, M.; Otani, M. Plant regeneration and Agrobacterium-mediated genetic transformation systems in liliaceous ornamental plants. Plant Biotechnol. 2020, 37, 129–140. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, X.; Zhang, H.; Li, S.; Li, Z. Establishment of a highly efficient in vitro propagation system of Diospyros lotus. Forests 2023, 14, 366. [Google Scholar] [CrossRef]
- Yasmin, A.; Debener, T. Transient gene expression in rose petals via Agrobacterium infiltration. Plant Cell Tissue Organ Cult. 2010, 102, 245–250. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, C.X.; Zheng, C.C.; Wen, F.J. Optimization of factors affecting genetic transformation of potato via Agrobacterium tumefaciens. J. Shandong Agric. Univ. (Nat. Sci.) 2002, 33, 23–27. [Google Scholar]
- Sutradhar, M.; Mandal, N. Reasons and riddance of Agrobacterium tumefaciens overgrowth in plant transformation. Transgenic Res. 2023, 32, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, X.; Li, B.; Lu, X.; Kang, J.; Cao, X. Establishment of in vitro culture system for Codonopsis pilosula transgenic hairy roots. 3 Biotech 2020, 10, 137. [Google Scholar] [CrossRef] [PubMed]
- Shinoyama, H.; Mitsuhara, I.; Ichikawa, H.; Kato, K.; Mochizuki, A. Transgenic chrysanthemums (Chrysanthemum morifolium Ramat.) carrying both insect and disease resistance. Acta Hortic. 2015, 1087, 485–497. [Google Scholar] [CrossRef]
- Naing, A.H.; Ai, T.N.; Jeon, S.M.; Lim, S.H.; Kim, C.K. An efficient protocol for Agrobacterium-mediated genetic transformation of recalcitrant chrysanthemum cultivar Shinma. Acta Physiol. Plant 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Firsov, A.; Mitiouchkina, T.; Shaloiko, L.; Pushin, A.; Vainstein, A.; Dolgov, S. Agrobacterium-mediated transformation of chrysanthemum with artemisinin biosynthesis pathway genes. Plants 2020, 9, 537. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, D.; Liu, Y.; Han, F.; Li, Z. A highly efficient genetic transformation system for broccoli and subcellular localization. Front. Plant Sci. 2023, 14, 1091588. [Google Scholar] [CrossRef]
- Song, S.; Yan, R.; Wang, C.; Wang, J.; Sun, H. Improvement of a genetic transformation system and preliminary study on the function of LpABCB21 and LpPILS7 based on somatic embryogenesis in Lilium pumilum DC. Fisch. Int. J. Mol. Sci. 2020, 21, 6784. [Google Scholar] [CrossRef]





| Recipient Explants | Total Number of Explants | GUS Positive Explants | GUS Transient Expression Rate |
|---|---|---|---|
| Petiole | 44 | 25 | 56.82% |
| Leaf | 40 | 5 | 12.5% |
| Callus | 45 | 7 | 15.56% |
| Treatment | Pre-Culture Time (d) | Co-Culture Time (d) | Bacterial Concentration (OD600) | Infection Time (min) | AS Concentration (μM) | Total Transformation Rate (%) |
|---|---|---|---|---|---|---|
| 2 | 3 | 3 | 0.6 | 11 | 200 | 9.88 |
| 7 | 3 | 3 | 0.8 | 15 | 200 | 2.36 |
| 18 | 3 | 3 | 0.4 | 11 | 200 | 1.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Tuo, W.; Wang, X.; Feng, B.; Xu, X.; Ahmad, S.; Zhai, J.; Peng, D.; Wu, S. Establishment of a Rapid and Effective Agrobacterium-Mediated Genetic Transformation System of Oxalis triangularis ‘Purpurea’. Plants 2023, 12, 4130. https://doi.org/10.3390/plants12244130
Xiao Y, Tuo W, Wang X, Feng B, Xu X, Ahmad S, Zhai J, Peng D, Wu S. Establishment of a Rapid and Effective Agrobacterium-Mediated Genetic Transformation System of Oxalis triangularis ‘Purpurea’. Plants. 2023; 12(24):4130. https://doi.org/10.3390/plants12244130
Chicago/Turabian StyleXiao, Yun, Wanli Tuo, Xuexuan Wang, Baomin Feng, Xinyu Xu, Sagheer Ahmad, Junwen Zhai, Donghui Peng, and Shasha Wu. 2023. "Establishment of a Rapid and Effective Agrobacterium-Mediated Genetic Transformation System of Oxalis triangularis ‘Purpurea’" Plants 12, no. 24: 4130. https://doi.org/10.3390/plants12244130





