In Vivo Phosphorylation of the Cytosolic Glucose-6-Phosphate Dehydrogenase Isozyme G6PD6 in Phosphate-Resupplied Arabidopsis thaliana Suspension Cells and Seedlings
Abstract
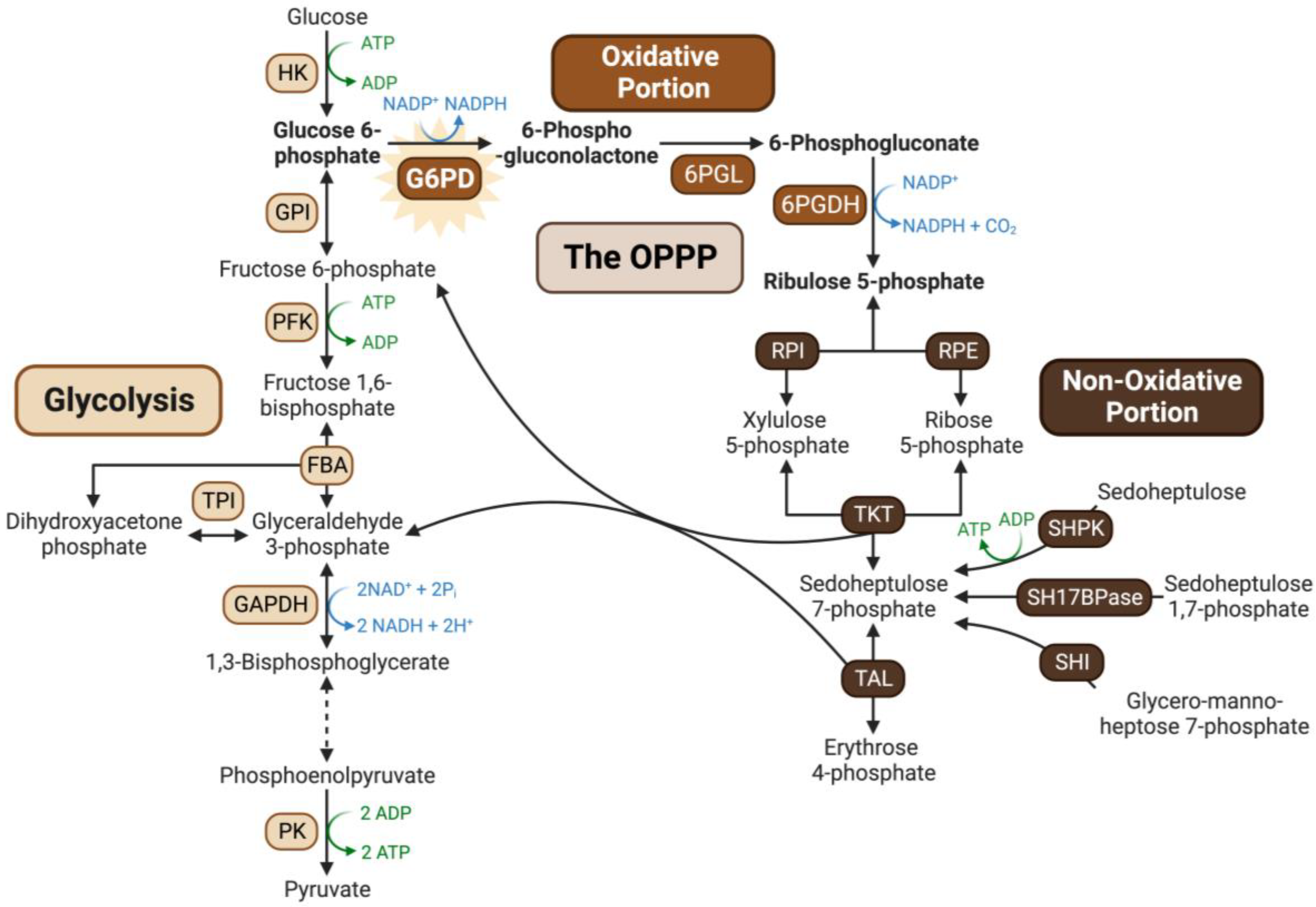
:1. Introduction
2. Results
2.1. Multisite N-Terminal Phosphorylation of Arabidopsis G6PD6 and Its Orthologs Is Quite Prevalent
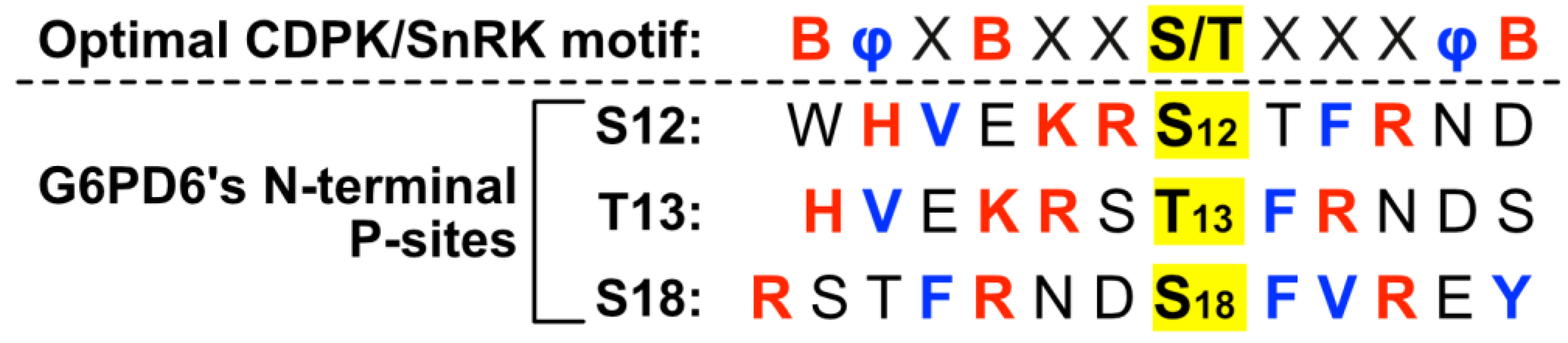
2.2. Residues Flanking G6PD6’s N-Terminal Phosphosites Are Representative of a Basophilic Motif
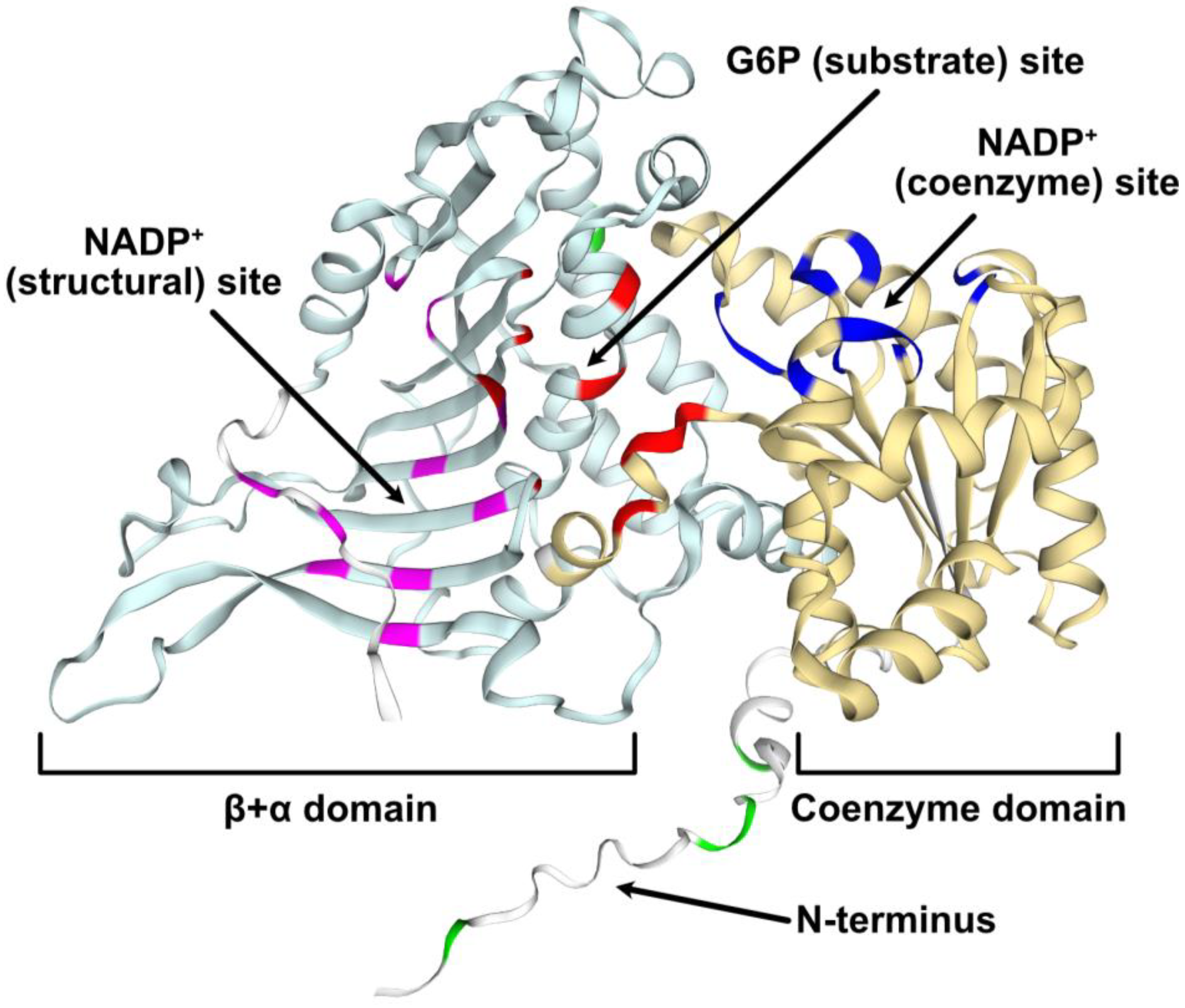
2.3. Structural Modeling of G6PD6 Indicates That Its N-Terminal Region Is Intrinsically Disordered
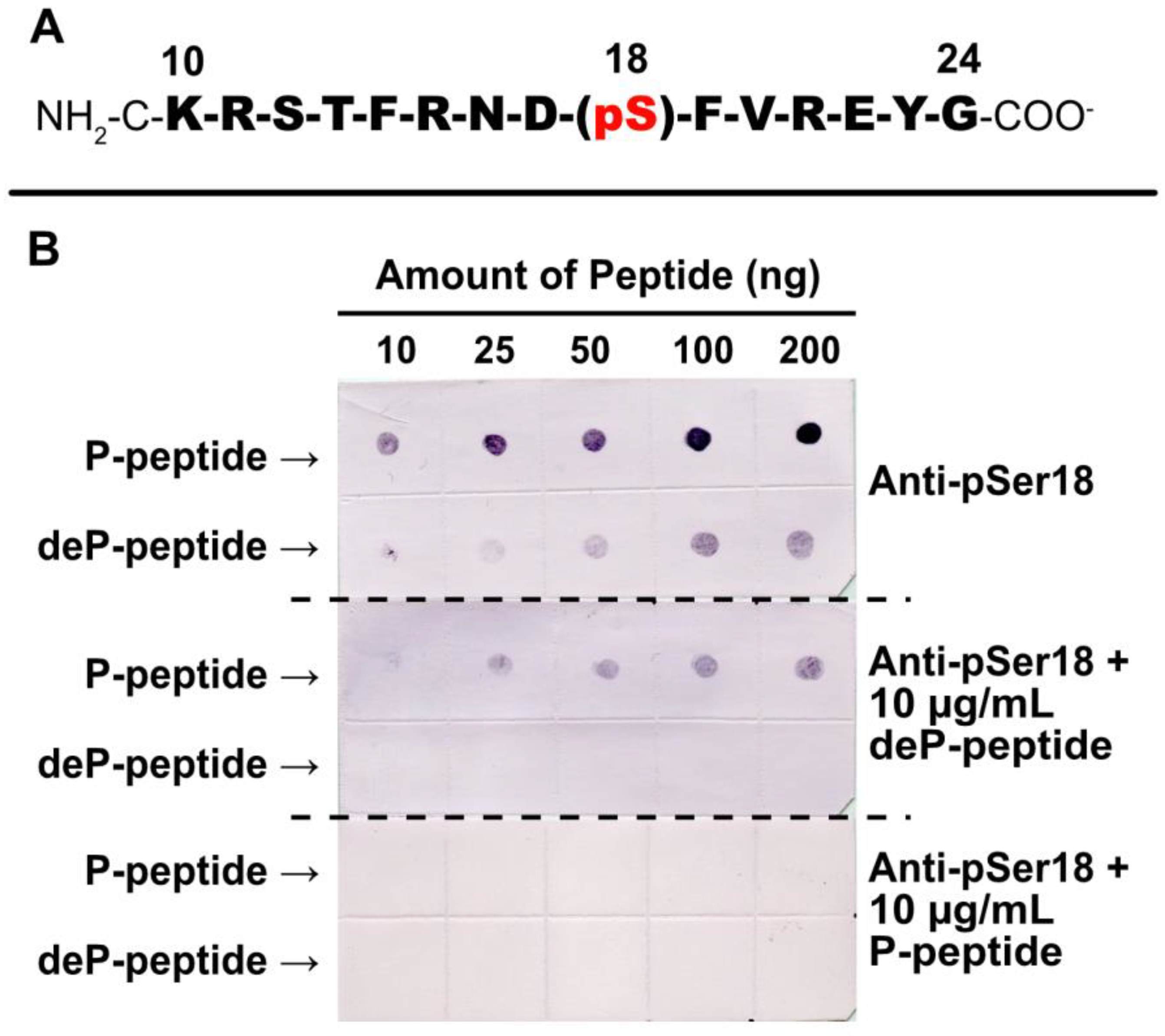
2.4. Specificity of Anti-pSer18 Immune Serum
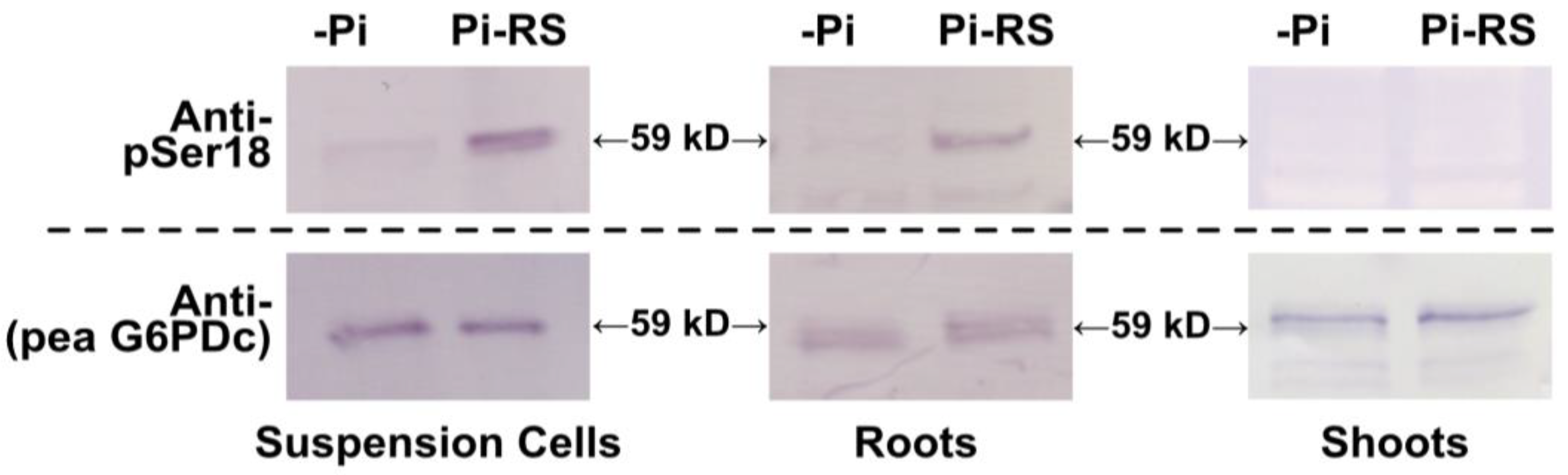
2.5. G6PD6 Phosphorylation at Ser18 Is Influenced by Pi Nutrition in Arabidopsis Suspension Cells and Roots
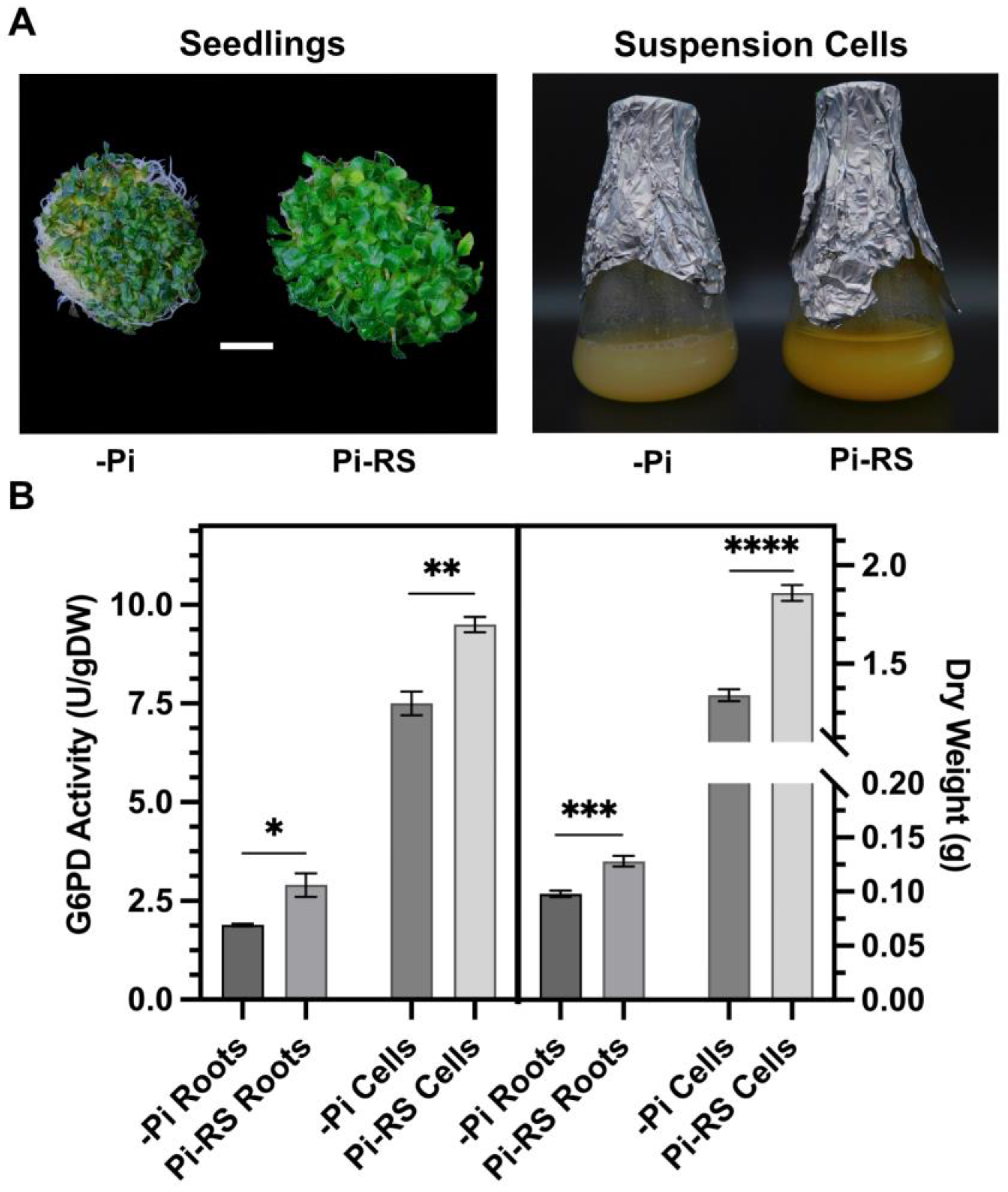
2.6. Pi-Resupply-Mediated Phosphorylation of Arabidopsis Root and Cell Culture G6PD6 at Ser18 Is Correlated with a Significant Increase in G6PD Activity and Biomass
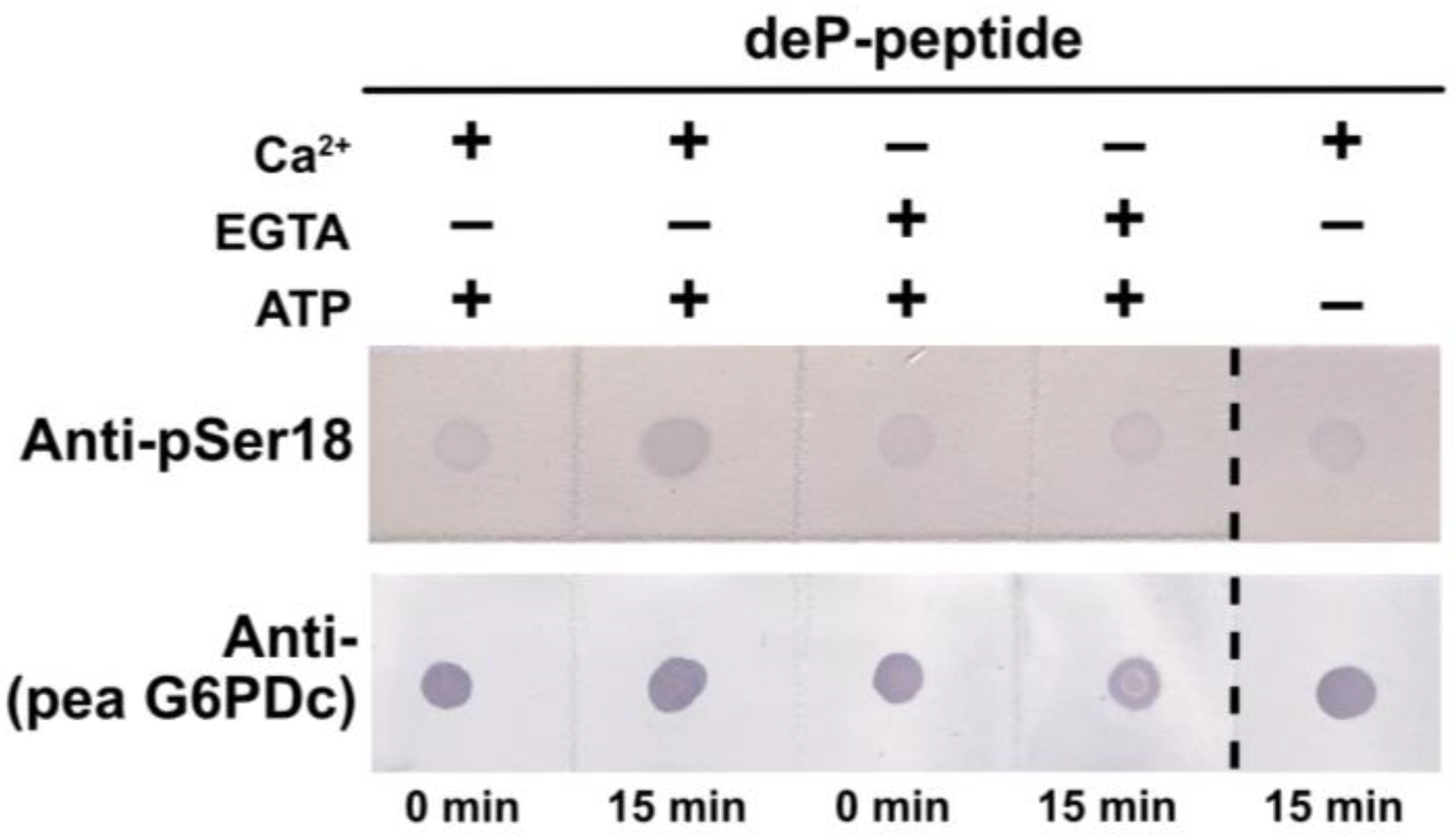
2.7. G6PD6 Phosphorylation at Ser18 Appears to Be Catalyzed by a CDPK
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Preparation of Clarified Protein Extracts
4.3. G6PD Activity Assays and Protein Concentration Determination
4.4. Preparation of Anti-(phosphoSer18-Specific AtG6PD6) Antibody
4.5. Electrophoresis and Immunoblotting
4.6. G6PD Peptide Kinase Assays
4.7. Bioinformatic Analyses
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, Z.; Wang, M.; Nicolas, M.; Ogé, L.; Pérez-Garcia, M.-D.; Crespel, L.; Li, G.; Ding, Y.; Le Gourrierec, J.; Grappin, P.; et al. Glucose-6-Phosphate Dehydrogenases: The Hidden Players of Plant Physiology. Int. J. Mol. Sci. 2022, 23, 16128. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Carfagna, S.; Massaro, G.; Vona, V.; Di Martino Rigano, V. Glucose-6-phosphate dehydrogenase in barley roots: Kinetic properties and localisation of the isoforms. Planta 2001, 212, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Massaro, G.; Vona, V.; Di Martino Rigano, V.; Carfagna, S.; Rigano, C. Ammonium induction of a novel isoform of glucose-6P dehydrogenase in barley roots. Physiol. Plant. 2001, 113, 469–476. [Google Scholar] [CrossRef]
- Wakao, S.; Benning, C. Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J. 2005, 41, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Castiglia, D.; Cardi, M.; Landi, S.; Cafasso, D.; Esposito, S. Expression and characterization of a cytosolic glucose 6 phosphate dehydrogenase isoform from barley (Hordeum vulgare) roots. Protein Expr. Purif. 2015, 112, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Linnenbrügger, L.; Doering, L.; Lansing, H.; Fischer, K.; Eirich, J.; Finkemeier, I.; von Schaewen, A. Alternative splicing of Arabidopsis G6PD5 recruits NADPH-producing OPPP reactions to the endoplasmic reticulum. Front. Plant Sci. 2022, 13, 909624. [Google Scholar] [CrossRef] [PubMed]
- Cardi, M.; Castiglia, D.; Ferrara, M.; Guerriero, G.; Chiurazzi, M.; Esposito, S. The effects of salt stress cause a diversion of basal metabolism in barley roots: Possible different roles for glucose-6-phosphate dehydrogenase isoforms. Plant Physiol. Biochem. 2015, 86, 44–54. [Google Scholar] [CrossRef]
- Dal Santo, S.; Stampfl, H.; Krasensky, J.; Kempa, S.; Gibon, Y.; Petutschnig, E.; Rozhon, W.; Heuck, A.; Clausen, T.; Jonak, C. Stress-Induced GSK3 Regulates the Redox Stress Response by Phosphorylating Glucose-6-Phosphate Dehydrogenase in Arabidopsis. Plant Cell 2012, 24, 3380–3392. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, Y.; Huang, S.; Yu, J.; Wang, X.; Xin, D.; Li, X.; Liu, Y.; Dai, Y.; Qi, Z.; et al. Genome-Wide Analysis of the Glucose-6-Phosphate Dehydrogenase Family in Soybean and Functional Identification of GmG6PDH2 Involvement in Salt Stress. Front. Plant Sci. 2020, 11, 214. [Google Scholar] [CrossRef]
- Nemoto, Y.; Sasakuma, T. Specific expression of glucose-6-phosphate dehydrogenase (G6PDH) gene by salt stress in wheat (Triticum aestivum L.). Plant Sci. 2000, 158, 53–60. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; Chang, N.; Nan, W.; Wang, S.; Ruan, M.; Sun, L.; Li, S.; Bi, Y. Cytosolic Glucose-6-Phosphate Dehydrogenase Is Involved in Seed Germination and Root Growth Under Salinity in Arabidopsis. Front. Plant Sci. 2019, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Wakao, S.; Andre, C.; Benning, C. Functional Analyses of Cytosolic Glucose-6-Phosphate Dehydrogenases and Their Contribution to Seed Oil Accumulation in Arabidopsis. Plant Physiol. 2007, 146, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Shane, M.W. The role of post-translational enzyme modifications in the metabolic adaptations of phosphorus-deprived plants. In Annual Plant Reviews; Roberts, J.A., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2015; Volume 48, pp. 99–124. [Google Scholar]
- Dissanayaka, D.M.S.B.; Ghahremani, M.; Siebers, M.; Wasaki, J.; Plaxton, W.C. Recent insights into the metabolic adaptations of phosphorus-deprived plants. J. Exp. Bot. 2021, 72, 199–223. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.L.; Hurley, B.A.; Tran, H.T.; Valentine, A.J.; She, Y.-M.; Knowles, V.L.; Plaxton, W.C. In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem. J. 2009, 420, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Shane, M.W.; Fedosejevs, E.T.; Plaxton, W.C. Reciprocal Control of Anaplerotic Phosphoenolpyruvate Carboxylase by in Vivo Monoubiquitination and Phosphorylation in Developing Proteoid Roots of Phosphate-Deficient Harsh Hakea. Plant Physiol. 2013, 161, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Masakapalli, S.K.; Bryant, F.M.; Kruger, N.J.; Ratcliffe, R.G. The metabolic flux phenotype of heterotrophic Arabidopsis cells reveals a flexible balance between the cytosolic and plastidic contributions to carbohydrate oxidation in response to phosphate limitation. Plant J. 2014, 78, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Ghahremani, M.; Pérez-Fernández, M.; Tan, M.; Schläpfer, P.; Plaxton, W.C.; Uhrig, R.G. Phosphate and phosphite have a differential impact on the proteome and phosphoproteome of Arabidopsis suspension cell cultures. Plant J. 2021, 105, 924–941. [Google Scholar] [CrossRef]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.-J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.-K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and Phosphoproteomics Reveal a Protein Phosphorylation Network in the Abscisic Acid Signaling Pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Z.; You, C.; Zhang, X.; Lu, J.; Ma, H. Abundant protein phosphorylation potentially regulates Arabidopsis anther development. J. Exp. Bot. 2016, 67, 4993–5008. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, H.; Berke, L.; Heck, A.J.R.; Mohammed, S.; Scheres, B.; Menke, F.L.H. Quantitative Phosphoproteomics after Auxin-stimulated Lateral Root Induction Identifies an SNX1 Protein Phosphorylation Site Required for Growth. Mol. Cell. Proteom. 2013, 12, 1158–1169. [Google Scholar] [CrossRef] [PubMed]
- Reiland, S.; Messerli, G.; Baerenfaller, K.; Gerrits, B.; Endler, A.; Grossmann, J.; Gruissem, W.; Baginsky, S. Large-Scale Arabidopsis Phosphoproteome Profiling Reveals Novel Chloroplast Kinase Substrates and Phosphorylation Networks. Plant Physiol. 2009, 150, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, G.B.; Wen, T.-N.; Nguyen, T.T.; Verslues, P.E. Protein Phosphatase 2Cs and Microtubule-Associated Stress Protein 1 Control Microtubule Stability, Plant Growth, and Drought Response. Plant Cell 2017, 29, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Roitinger, E.; Hofer, M.; Köcher, T.; Pichler, P.; Novatchkova, M.; Yang, J.; Schlögelhofer, P.; Mechtler, K. Quantitative Phosphoproteomics of the Ataxia Telangiectasia-Mutated (ATM) and Ataxia Telangiectasia-Mutated and Rad3-related (ATR) Dependent DNA Damage Response in Arabidopsis thaliana. Mol. Cell. Proteom. 2015, 14, 556–571. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Wang, P.; Wang, L.; Renzi, E.; Radivojac, P.; Tang, H.; Arnold, R.; Zhu, J.-K.; Tao, W.A. Quantitative Measurement of Phosphoproteome Response to Osmotic Stress in Arabidopsis Based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol. Cell. Proteom. 2013, 12, 2354–2369. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Brachova, L.; Nikolau, B.J.; Jones, A.M.; Walley, J.W. Heterotrimeric G-Protein-Dependent Proteome and Phosphoproteome in Unstimulated Arabidopsis Roots. Proteomics 2018, 18, e1800323. [Google Scholar] [CrossRef]
- Reiland, S.; Finazzi, G.; Endler, A.; Willig, A.; Baerenfaller, K.; Grossmann, J.; Gerrits, B.; Rutishauser, D.; Gruissem, W.; Rochaix, J.-D.; et al. Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc. Natl. Acad. Sci. USA 2011, 108, 12955–12960. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, B.; Née, G.; Kramer, K.; Finkemeier, I.; Soppe, W.J.J. Sequence Polymorphisms at the REDUCED DORMANCY5 Pseudophosphatase Underlie Natural Variation in Arabidopsis Dormancy. Plant Physiol. 2016, 171, 2659–2670. [Google Scholar] [CrossRef]
- Engelsberger, W.R.; Schulze, W.X. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 2012, 69, 978–995. [Google Scholar] [CrossRef]
- Wang, X.; Bian, Y.; Cheng, K.; Gu, L.-F.; Ye, M.; Zou, H.; Sun, S.S.-M.; He, J.-X. A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J. Proteom. 2013, 78, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Wei, Y.; Xie, Y.; Liu, L.; Jiang, C.; Yu, Y. Quantitative phosphoproteomic analysis provides insights into the aluminum-responsiveness of Tamba black soybean. PLoS ONE 2020, 15, e0237845. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.; Yao, L.; Ma, Z.; Ren, P.; Si, E.; Li, B.; Meng, Y.; Ma, X.; Yang, K.; et al. Global proteome analyses of phosphorylation and succinylation of barley root proteins in response to phosphate starvation and recovery. Front. Plant Sci. 2022, 13, 917652. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Qiu, J.; Wang, Y.; Li, Z.; Zhao, J.; Tong, X.; Lin, H.; Zhang, J. A Quantitative Proteomic Analysis of Brassinosteroid-induced Protein Phosphorylation in Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 514. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Hou, Y.; Tong, X.; Wang, Y.; Lin, H.; Liu, Q.; Zhang, W.; Li, Z.; Nallamilli, B.R.; Zhang, J. Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tong, X.; Qiu, J.; Li, Z.; Zhao, J.; Hou, Y.; Tang, L.; Zhang, J. A phosphoproteomic landscape of rice (Oryza sativa) tissues. Physiol. Plant. 2017, 160, 458–475. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-Scale Comparative Phosphoproteomics Identifies Conserved Phosphorylation Sites in Plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef]
- Qiu, J.; Hou, Y.; Wang, Y.; Li, Z.; Zhao, J.; Tong, X.; Lin, H.; Wei, X.; Ao, H.; Zhang, J. A Comprehensive Proteomic Survey of ABA-Induced Protein Phosphorylation in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2017, 18, 60. [Google Scholar] [CrossRef]
- Xi, L.; Zhang, Z.; Herold, S.; Kassem, S.; Wu, X.N.; Schulze, W.X. Phosphorylation site motifs in plant protein kinases and their substrates. In Plant Phosphoproteomics: Methods and Protocols; Wu, X.N., Ed.; Springer: New York, NY, USA, 2021; pp. 1–16. [Google Scholar]
- Huang, J.-Z.; Huber, S.C. Phosphorylation of Synthetic Peptides by a CDPK and Plant SNF1-Related Protein Kinase. Influence of Proline and Basic Amino Acid Residues at Selected Positions. Plant Cell Physiol. 2001, 42, 1079–1087. [Google Scholar] [CrossRef]
- Harper, J.F.; Harmon, A. Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 2005, 6, 555–566. [Google Scholar] [CrossRef]
- Kotaka, M.; Gover, S.; Vandeputte-Rutten, L.; Au, S.W.N.; Lam, V.M.S.; Adams, M.J. Structural studies of glucose-6-phosphate and NADP+binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr. Sect. D Struct. Biol. 2005, 61, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Fickenscher, K.; Scheibe, R. Purification and properties of the cytoplasmic glucose-6-phosphate dehydrogenase from pea leaves. Arch. Biochem. Biophys. 1986, 247, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Lendzian, K.J. Modulation of glucose-6-phosphate dehydrogenase by NADPH, NADP+ and dithiothreitol at variable NADPH/NADP+ ratios in an illuminated reconstituted Spinach (Spinacia oleracea L.) chloroplast system. Planta 1980, 148, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Douglas, P.; Moorhead, G.; Hong, Y.; Morrice, N.; MacKintosh, C. Purification of a nitrate reductase kinase from Spinacea oleracea leaves, and its identification as a calmodulin-domain protein kinase. Planta 1998, 206, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Fedosejevs, E.T.; Gerdis, S.A.; Ying, S.; Pyc, M.; Anderson, E.M.; Snedden, W.A.; Mullen, R.T.; She, Y.-M.; Plaxton, W.C. The calcium-dependent protein kinase RcCDPK2 phosphorylates sucrose synthase at Ser11 in developing castor oil seeds. Biochem. J. 2016, 473, 3667–3682. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Hill, A.T.; Pyc, M.; Anderson, E.M.; Snedden, W.A.; Mullen, R.T.; She, Y.-M.; Plaxton, W.C. Regulatory Phosphorylation of Bacterial-Type PEP Carboxylase by the Ca2+-Dependent Protein Kinase RcCDPK1 in Developing Castor Oil Seeds. Plant Physiol. 2017, 174, 1012–1027. [Google Scholar] [CrossRef]
- Liu, K.-H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Chung, H.S.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Au, S.W.; Gover, S.; Lam, V.M.; Adams, M.J. Human glucose-6-phosphate dehydrogenase: The crystal structure reveals a structural NADP + molecule and provides insights into enzyme deficiency. Structure 2000, 8, 293–303. [Google Scholar] [CrossRef]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef]
- Park, J.; Khuu, N.; Howard, A.S.M.; Mullen, R.T.; Plaxton, W.C. Bacterial- and plant-type phosphoenolpyruvate carboxylase isozymes from developing castor oil seeds interact in vivo and associate with the surface of mitochondria. Plant J. 2012, 71, 251–262. [Google Scholar] [CrossRef]
- O’Leary, B.; Rao, S.K.; Plaxton, W.C. Phosphorylation of bacterial-type phosphoenolpyruvate carboxylase at Ser425 provides a further tier of enzyme control in developing castor oil seeds. Biochem. J. 2011, 433, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Dalziel, K.J.; O’Leary, B.; Brikis, C.; Rao, S.K.; She, Y.-M.; Cyr, T.; Plaxton, W.C. The bacterial-type phosphoenolpyruvate carboxylase isozyme from developing castor oil seeds is subject to in vivo regulatory phosphorylation at serine-451. FEBS Lett. 2012, 586, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Moorhead, G.B.G.; Templeton, G.W.; Tran, H.T. Role of protein kinases, phosphatases and 14-3-3 proteins in the control of primary plant metabolism. In Annual Plant Reviews Volume 22: Control of Primary Metabolism in Plants; Plaxton, W.C., McManus, M.T., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 121–143. [Google Scholar]
- Brumbaugh, K.; Johnson, W.; Liao, W.-C.; Lin, M.-S.; Houchins, J.P.; Cooper, J.; Stoesz, S.; Campos-Gonzalez, R. Overview of the generation, validation, and application of phosphosite-specific antibodies. In Signal Transduction Immunohistochemistry: Methods and Protocols; Kalyuzhny, A.E., Ed.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2011; Volume 717, pp. 3–43. [Google Scholar]
- Hartman, M.D.; Rojas, B.E.; Ferrero, D.M.; Leyva, A.; Durán, R.; Iglesias, A.A.; Figueroa, C.M. Phosphorylation of aldose-6-phosphate reductase from Prunus persica leaves. Plant Physiol. Biochem. 2023, 194, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Fedosejevs, E.T.; Ying, S.; Park, J.; Anderson, E.M.; Mullen, R.T.; She, Y.-M.; Plaxton, W.C. Biochemical and Molecular Characterization of RcSUS1, a Cytosolic Sucrose Synthase Phosphorylated in Vivo at Serine 11 in Developing Castor Oil Seeds. J. Biol. Chem. 2014, 289, 33412–33424. [Google Scholar] [CrossRef]
- Huber, S.C.; Huber, J.L. Role and regulation of sucrose-phosphate synthase in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 431–444. [Google Scholar] [CrossRef]
- Stampfl, H.; Fritz, M.; Dal Santo, S.; Jonak, C. The GSK3/Shaggy-like kinase ASKα contributes to pattern-triggered immunity in Arabidopsis thaliana. Plant Physiol. 2016, 171, 1366–1377. [Google Scholar] [CrossRef]
- Veljanovski, V.; Vanderbeld, B.; Knowles, V.L.; Snedden, W.A.; Plaxton, W.C. Biochemical and Molecular Characterization of AtPAP26, a Vacuolar Purple Acid Phosphatase Up-Regulated in Phosphate-Deprived Arabidopsis Suspension Cells and Seedlings. Plant Physiol. 2006, 142, 1282–1293. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Penefsky, H.S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 1977, 252, 2891–2899. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Lin, S.; Wang, C.; Zhou, J.; Shi, Y.; Ruan, C.; Tu, Y.; Yao, L.; Peng, D.; Xue, Y. EPSD: A well-annotated data resource of protein phosphorylation sites in eukaryotes. Brief. Bioinform. 2020, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.; Horne, A.; Van Parys, T.; Goormachtig, S.; De Smet, I.; Botzki, A.; Van Breusegem, F.; Gevaert, K. The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J. 2019, 99, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, A.G.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2022, 51, D418–D427. [Google Scholar] [CrossRef]
- Piovesan, D.; Del Conte, A.; Clementel, D.; Monzon, A.M.; Bevilacqua, M.; Aspromonte, M.C.; Iserte, A.J.; Orti, E.F.; Marino-Buslje, C.; Tosatto, S.C.E. MobiDB: 10 years of intrinsically disordered proteins. Nucleic Acids Res. 2023, 51, D438–D444. [Google Scholar] [CrossRef]

| Species | G6PDc Isozyme | Condition | Tissue | Phosphosites | Reference |
|---|---|---|---|---|---|
| Arabidopsis thaliana | G6PD6 | Abscisic acid (ABA) application | Seedling | S3, T13, S18 | [19,20] |
| Development | Anther | T13, S18 | [21] | ||
| Auxin application | Root | S18 | [22] | ||
| Day-night transition | Rosette | S18 | [23] | ||
| Drought | Seedling | S3, T13, S18 | [20,24] | ||
| Ionizing radiation | Seedling | T13, S18 | [25] | ||
| Mannitol | Seedling | S18 | [26] | ||
| Pi resupply | Cell culture | S12, T13, S18 | [18] | ||
| Unstressed | Multiple | S3, S12, T13, S18 | [27,28,29] | ||
| G6PD5 | Seed imbibition | Seed | S3 | [30] | |
| Nitrogen re-supply | Seedling | S488 | [31] | ||
| Unstressed | Multiple | S3, S12, T13, S18 | [27,32] | ||
| Glycine max | G6PD4 | Aluminum stress | Root | S18 | [33] |
| Hordeum vulgare | G6PD | Pi resupply | Root | S14, Y395 | [34] |
| Oryza sativa | G6PD1 | Brassinosteroid application | Seedling | S16, S19, S21 | [35] |
| Seed development | Seed | S21 | [36] | ||
| Multiple | S16, S21, Y501, T504, T513, S515 | [37,38] | |||
| G6PD2 | ABA application | Seedling | S13 | [39] | |
| Brassinosteroid application | Seedling | S13 | [35] | ||
| Seed development | Seed | S13 | [36] | ||
| Unstressed | Multiple | S12, S13, S16 | [37,38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, M.A.; Benidickson, K.H.; Plaxton, W.C. In Vivo Phosphorylation of the Cytosolic Glucose-6-Phosphate Dehydrogenase Isozyme G6PD6 in Phosphate-Resupplied Arabidopsis thaliana Suspension Cells and Seedlings. Plants 2024, 13, 31. https://doi.org/10.3390/plants13010031
Smith MA, Benidickson KH, Plaxton WC. In Vivo Phosphorylation of the Cytosolic Glucose-6-Phosphate Dehydrogenase Isozyme G6PD6 in Phosphate-Resupplied Arabidopsis thaliana Suspension Cells and Seedlings. Plants. 2024; 13(1):31. https://doi.org/10.3390/plants13010031
Chicago/Turabian StyleSmith, Milena A., Kirsten H. Benidickson, and William C. Plaxton. 2024. "In Vivo Phosphorylation of the Cytosolic Glucose-6-Phosphate Dehydrogenase Isozyme G6PD6 in Phosphate-Resupplied Arabidopsis thaliana Suspension Cells and Seedlings" Plants 13, no. 1: 31. https://doi.org/10.3390/plants13010031









