Identification and Characterization of Jasmonic Acid Methyltransferase Involved in the Formation of Floral Methyl Jasmonate in Hedychium coronarium
Abstract
:1. Introduction
2. Results
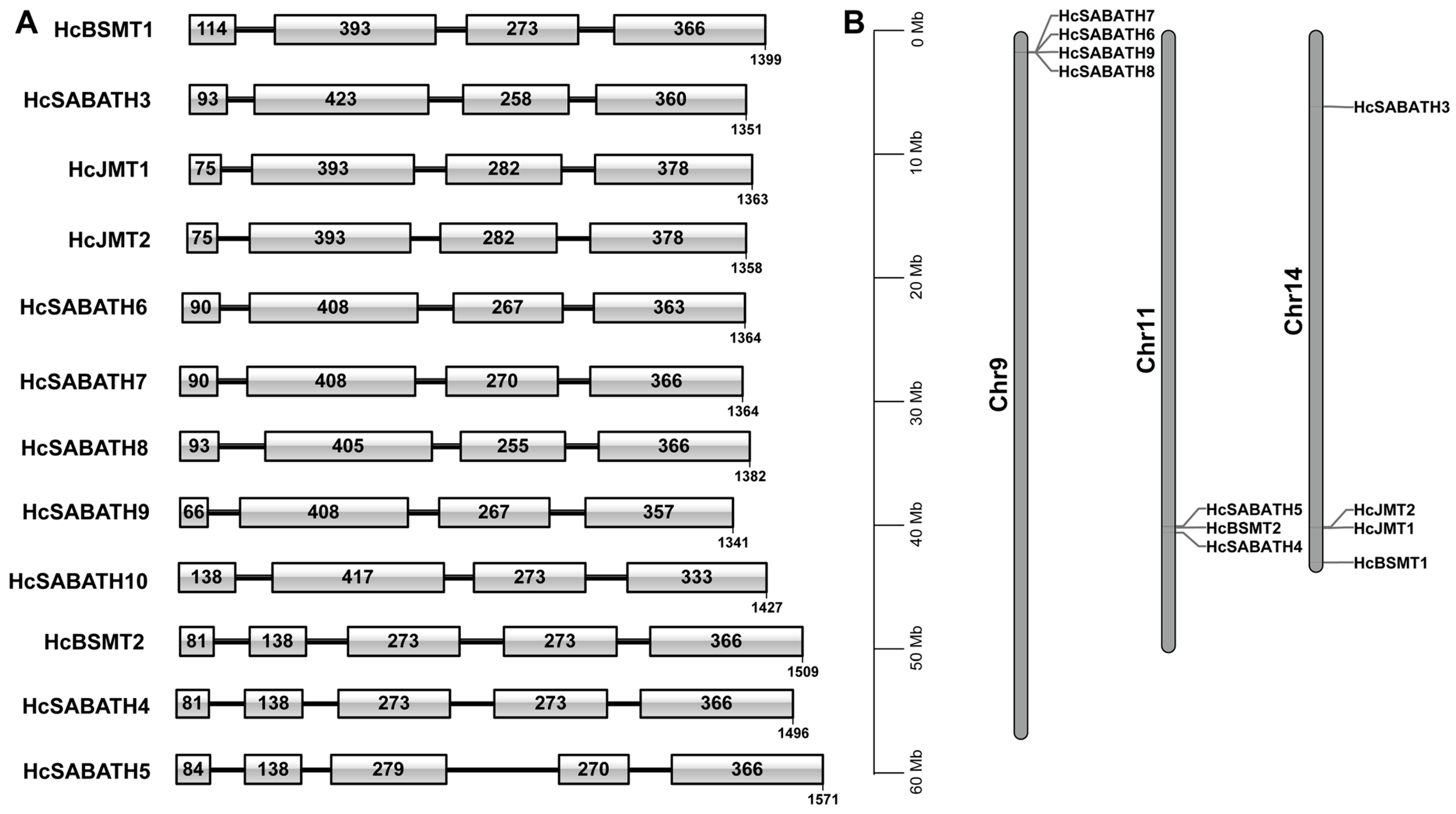
2.1. Identification of SABATH Gene Family Members in H. coronarium
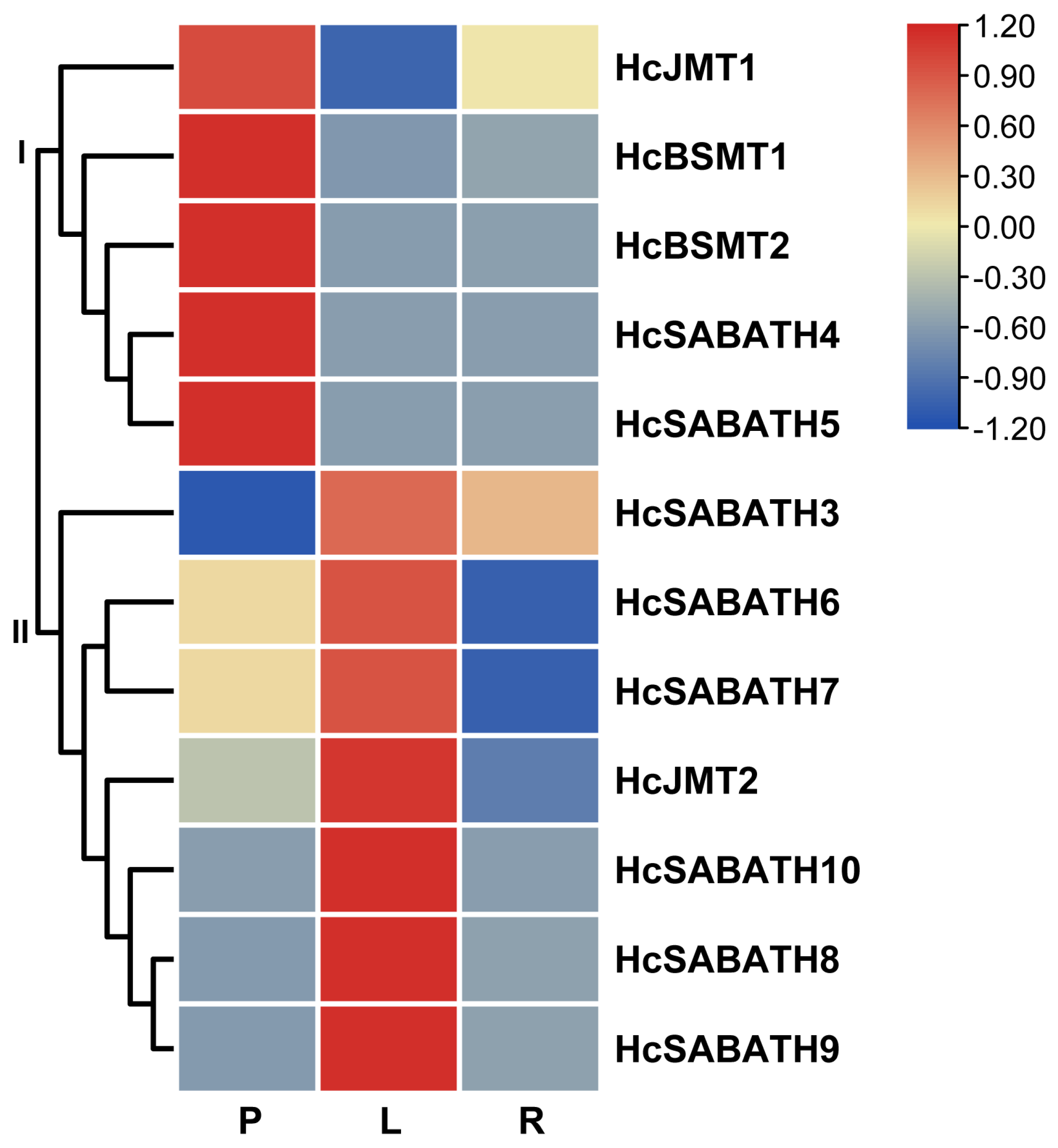
2.2. Phylogenetic and Expression Profiling Analyses Revealed HcJMT1 as a Candidate for Floral MeJA Formation
2.3. Cloning and Sequence Analysis of HcJMT1
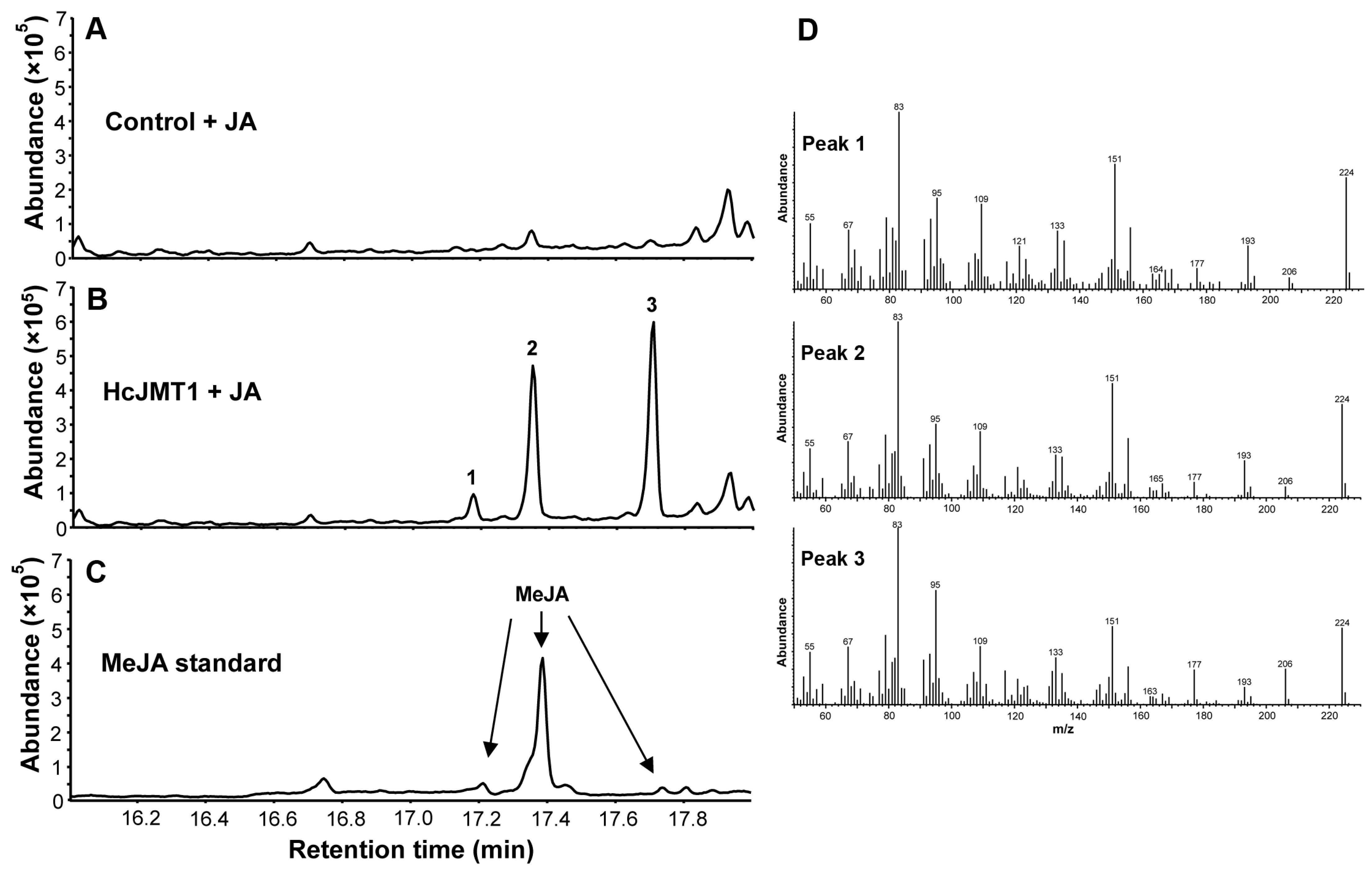
2.4. HcJMT1 Is a Jasmonic Acid Methyltransferase
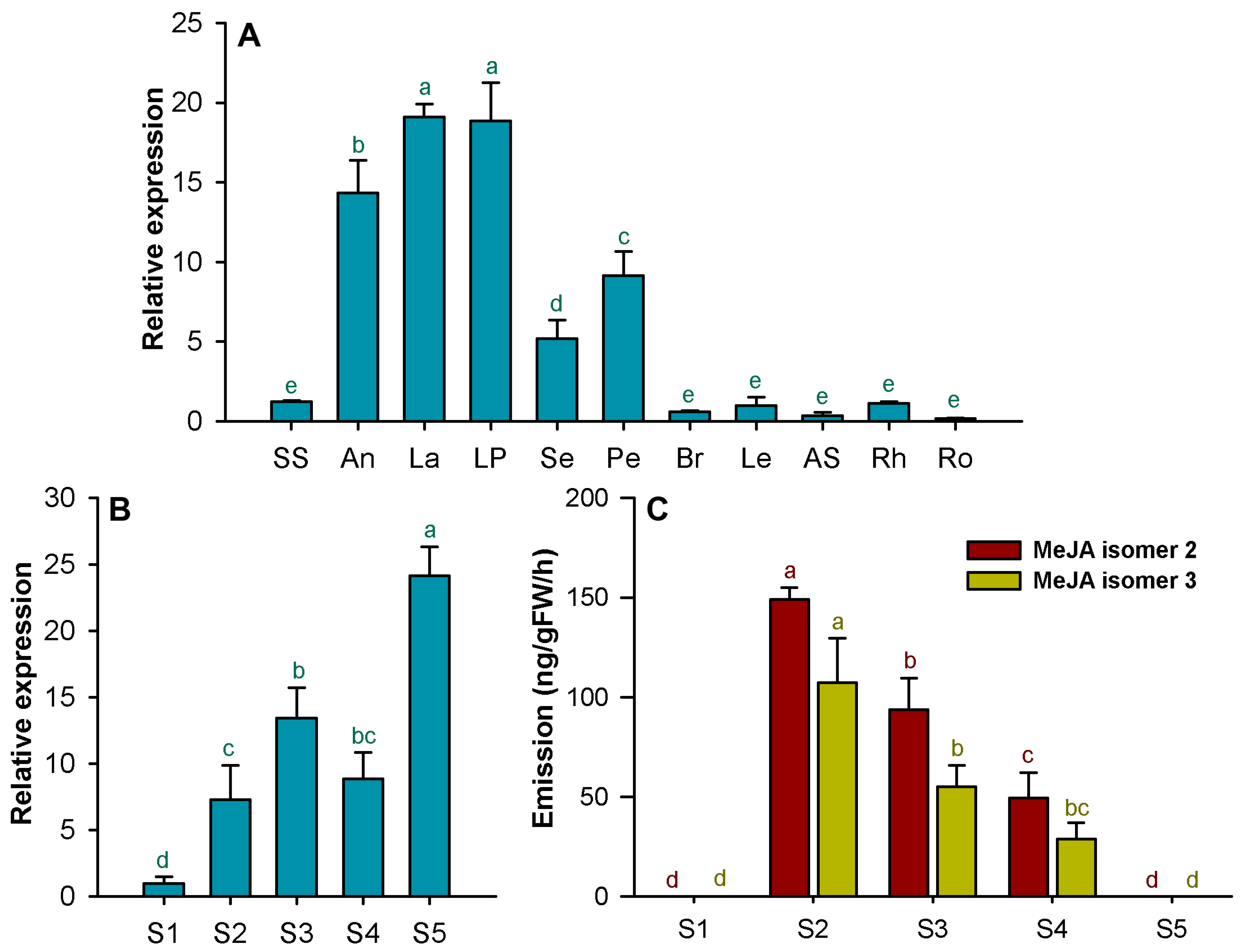
2.5. Expression Analysis of HcJMT1
3. Discussion
3.1. Evolution of the SABATH Methyltransferase Family in H. coronarium
3.2. Involvement of HcJMT1 in the Biosynthesis of Floral MeJA
3.3. Possible Roles of Floral MeJA in H. coronarium
4. Materials and Methods
4.1. Plant Materials
4.2. Genome-Wide Identification of SABATH Genes in H. coronarium
4.3. Phylogenetic Analysis and Expression Profiling of HcSABATH Genes
4.4. Isolation of the HcJMT1 Gene
4.5. Bacterial Expression and Purification of the HcJMT1 Recombinant Protein
4.6. In Vitro Enzyme Assays of HcJMT1
4.7. Real-Time PCR
4.8. Headspace Collection and GC-MS Analysis of Floral MeJA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muhlemann, J.K.; Klempien, A.; Dudareva, N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014, 37, 1936–1949. [Google Scholar] [CrossRef]
- Raguso, R.A. Wake up and smell the roses: The ecology and evolution of floral scent. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 549–569. [Google Scholar] [CrossRef]
- Bouwmeester, H.; Schuurink, R.C.; Bleeker, P.M.; Schiestl, F. The role of volatiles in plant communication. Plant J. 2019, 100, 892–907. [Google Scholar] [CrossRef]
- Ninkovic, V.; Markovic, D.; Rensing, M. Plant volatiles as cues and signals in plant communication. Plant Cell Environ. 2021, 44, 1030–1043. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Yue, Y.; Wang, L.; Li, M.; Liu, F.; Yin, J.; Huang, L.; Zhou, B.; Li, X.; Yu, Y.; Chen, F.; et al. A BAHD acyltransferase contributes to the biosynthesis of both ethyl benzoate and methyl benzoate in the flowers of Lilium oriental hybrid ‘Siberia’. Front. Plant Sci. 2023, 14, 1275960. [Google Scholar] [CrossRef]
- Bao, T.; Kimani, S.; Li, Y.; Li, H.; Yang, S.; Zhang, J.; Wang, Q.; Wang, Z.; Ning, G.; Wang, L.; et al. Allelic variation of terpene synthases drives terpene diversity in the wild species of the Freesia genus. Plant Physiol. 2023, 192, 2419–2435. [Google Scholar] [CrossRef]
- Knudsen, J.T.; Eriksson, R.; Gershenzon, J.; Stahl, B. Diversity and distribution of floral scent. Bot. Rev. 2006, 72, 1–120. [Google Scholar] [CrossRef]
- Cheong, J.J.; Choi, Y.D. Methyl jasmonate as a vital substance in plants. Trends Genet. 2003, 19, 409–413. [Google Scholar] [CrossRef]
- Ai, Y.; Li, Z.; Sun, W.H.; Chen, J.; Zhang, D.; Ma, L.; Zhang, Q.H.; Chen, M.K.; Zheng, Q.D.; Liu, J.F.; et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 255. [Google Scholar] [CrossRef]
- Tian, C.; Liu, S.; Jiang, L.; Tian, S.; Wang, G. The expression characteristics of methyl jasmonate biosynthesis-related genes in Cymbidium faberi and influence of heterologous expression of CfJMT in Petunia hybrida. Plant Physiol. Biochem. 2020, 151, 400–410. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, C.; Niu, T.; Chen, M.; Guo, L.; Hou, X. Identification of floral volatile components and expression analysis of controlling gene in Paeonia ostii ‘Fengdan’ under different cultivation conditions. Plants 2023, 12, 2453. [Google Scholar] [CrossRef]
- Zhou, Y.; Abbas, F.; Wang, Z.; Yu, Y.; Yue, Y.; Li, X.; Yu, R.; Fan, Y. HS-SPME-GC-MS and electronic nose reveal differences in the volatile profiles of Hedychium flowers. Molecules 2021, 26, 5425. [Google Scholar] [CrossRef]
- Huang, M.; Ma, C.; Yu, R.; Mu, L.; Hou, J.; Yu, Y.; Fan, Y. Concurrent changes in methyl jasmonate emission and the expression of its biosynthesis-related genes in Cymbidium ensifolium flowers. Physiol. Plant. 2015, 153, 503–512. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Qi, J.; Li, J.; Han, X.; Li, R.; Wu, J.; Yu, H.; Hu, L.; Xiao, Y.; Lu, J.; Lou, Y. Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 2016, 58, 564–576. [Google Scholar] [CrossRef]
- Seo, H.S.; Song, J.T.; Cheong, J.J.; Lee, Y.H.; Lee, Y.W.; Hwang, I.; Lee, J.S.; Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef]
- Zhao, N.; Yao, J.; Chaiprasongsuk, M.; Li, G.; Guan, J.; Tschaplinski, T.J.; Guo, H.; Chen, F. Molecular and biochemical characterization of the jasmonic acid methyltransferase gene from black cottonwood (Populus trichocarpa). Phytochemistry 2013, 94, 74–81. [Google Scholar] [CrossRef]
- Teng, D.; Jing, W.; Lv, B.; Huang, X.; Zhao, D.; Kou, J.; Liu, X.; Dhiloo, K.H.; Zhang, Y. Two jasmonic acid carboxyl methyltransferases in Gossypium hirsutum involved in MeJA biosynthesis may contribute to plant defense. Front. Plant Sci. 2023, 14, 1249226. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, H.; Zhu, B.; Li, J.; Yang, T.; Zhang, Z.Z.; Deng, W.W. Molecular and biochemical characterization of jasmonic acid carboxyl methyltransferase involved in aroma compound production of methyl jasmonate during black tea processing. J. Agric. Food Chem. 2021, 69, 3154–3164. [Google Scholar] [CrossRef]
- Preuss, A.; Augustin, C.; Figueroa, C.R.; Hoffmann, T.; Valpuesta, V.; Sevilla, J.F.; Schwab, W. Expression of a functional jasmonic acid carboxyl methyltransferase is negatively correlated with strawberry fruit development. J. Plant Physiol. 2014, 171, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C.; Chen, F.; Pichersky, E. The SABATH family of MTs in Arabidopsis thaliana and other plant species. In Recent Advances in Phytochemistry; Romeo, J., Ed.; Elsevier: Amsterdam, The Netherlands, 2003; Volume 37, pp. 253–283. [Google Scholar] [CrossRef]
- Qu, L.; Li, S.; Xing, S. Methylation of phytohormones by the SABATH methyltransferases. Chin. Sci. Bull. 2010, 55, 2211–2218. [Google Scholar] [CrossRef]
- Effmert, U.; Saschenbrecker, S.; Ross, J.; Negre, F.; Fraser, C.M.; Noel, J.P.; Dudareva, N.; Piechulla, B. Floral benzenoid carboxyl methyltransferases: From in vitro to in planta function. Phytochemistry 2005, 66, 1211–1230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, X.; Crandall-Stotler, B.; Qian, P.; Kollner, T.G.; Guo, H.; Chen, F. Biosynthesis of methyl (E)-cinnamate in the liverwort Conocephalum salebrosum and evolution of cinnamic acid methyltransferase. Phytochemistry 2019, 164, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Pillet, J.; Chambers, A.H.; Barbey, C.; Bao, Z.; Plotto, A.; Bai, J.; Schwieterman, M.; Johnson, T.; Harrison, B.; Whitaker, V.M.; et al. Identification of a methyltransferase catalyzing the final step of methyl anthranilate synthesis in cultivated strawberry. BMC Plant Biol. 2017, 17, 147. [Google Scholar] [CrossRef]
- Dudareva, N.; Raguso, R.A.; Wang, J.; Ross, J.R.; Pichersky, E. Floral scent production in Clarkia breweri. III. Enzymatic synthesis and emission of benzenoid esters. Plant Physiol. 1998, 116, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Murfitt, L.M.; Kolosova, N.; Mann, C.J.; Dudareva, N. Purification and characterization of S-adenosyl-L-methionine: Benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methyl benzoate in flowers of Antirrhinum majus. Arch. Biochem. Biophys. 2000, 382, 145–151. [Google Scholar] [CrossRef]
- Kato, M.; Mizuno, K.; Crozier, A.; Fujimura, T.; Ashihara, H. Caffeine synthase gene from tea leaves. Nature 2000, 406, 956–957. [Google Scholar] [CrossRef]
- Zhao, N.; Ferrer, J.L.; Moon, H.S.; Kapteyn, J.; Zhuang, X.; Hasebe, M.; Stewart, C.J.; Gang, D.R.; Chen, F. A SABATH methyltransferase from the moss Physcomitrella patens catalyzes S-methylation of thiols and has a role in detoxification. Phytochemistry 2012, 81, 31–41. [Google Scholar] [CrossRef]
- Chaiprasongsuk, M.; Zhang, C.; Qian, P.; Chen, X.; Li, G.; Trigiano, R.N.; Guo, H.; Chen, F. Biochemical characterization in Norway spruce (Picea abies) of SABATH methyltransferases that methylate phytohormones. Phytochemistry 2018, 149, 146–154. [Google Scholar] [CrossRef]
- Zhao, N.; Ferrer, J.; Ross, J.; Guan, J.; Yang, Y.; Pichersky, E.; Noel, J.P.; Chen, F. Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol. 2008, 146, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Tao, K.; Zhang, J.; Lu, S.; Chen, S.; Liao, J. Identification of SABATH family members in Solanum lycopersicum and their expression patterns under abiotic/biotic stresses. Plant Mol. Biol. Rep. 2021, 39, 403–418. [Google Scholar] [CrossRef]
- Guo, Y.; Qiao, D.; Yang, C.; Chen, J.; Li, Y.; Liang, S.; Lin, K.; Chen, Z. Genome-wide identification and expression analysis of SABATH methyltransferases in tea plant (Camellia sinensis): Insights into their roles in plant defense responses. Plant Signal. Behav. 2020, 15, 1804684. [Google Scholar] [CrossRef] [PubMed]
- Han, X.M.; Yang, Q.; Liu, Y.J.; Yang, Z.L.; Wang, X.R.; Zeng, Q.Y.; Yang, H.L. Evolution and function of the Populus SABATH family reveal that a single amino acid change results in a substrate switch. Plant Cell Physiol. 2018, 59, 392–403. [Google Scholar] [CrossRef]
- Yue, Y.; Wang, L.; Yu, R.; Chen, F.; He, J.; Li, X.; Yu, Y.; Fan, Y. Coordinated and high-level expression of biosynthetic pathway genes is responsible for the production of a major floral scent compound methyl benzoate in Hedychium coronarium. Front. Plant Sci. 2021, 12, 650582. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Yu, R.; Fan, Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 2014, 240, 745–762. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Yu, R.; Fan, Y. Transcriptome profiling provides new insights into the formation of floral scent in Hedychium coronarium. BMC Genom. 2015, 16, 470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chaiprasongsuk, M.; Chanderbali, A.S.; Chen, X.; Fu, J.; Soltis, D.E.; Chen, F. Origin and evolution of a gibberellin-deactivating enzyme GAMT. Plant Direct 2020, 4, e287. [Google Scholar] [CrossRef]
- Qin, G.; Gu, H.; Zhao, Y.; Ma, Z.; Shi, G.; Yang, Y.; Pichersky, E.; Chen, H.; Liu, M.; Chen, Z.; et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005, 17, 2693–2704. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, J.S.; Ross, J.; Noel, J.P.; Pichersky, E.; Chen, F. An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 2006, 448, 123–132. [Google Scholar] [CrossRef]
- Zubieta, C.; Ross, J.R.; Koscheski, P.; Yang, Y.; Pichersky, E.; Noel, J.P. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 2003, 15, 1704–1716. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef]
- Stitz, M.; Hartl, M.; Baldwin, I.T.; Gaquerel, E. Jasmonoyl-L-isoleucine coordinates metabolic networks required for anthesis and floral attractant emission in wild tobacco (Nicotiana attenuata). Plant Cell 2014, 26, 3964–3983. [Google Scholar] [CrossRef]
- Kim, E.H.; Kim, Y.S.; Park, S.H.; Koo, Y.J.; Choi, Y.D.; Chung, Y.Y.; Lee, I.J.; Kim, J.K. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009, 149, 1751–1760. [Google Scholar] [CrossRef]
- Chen, F.; D’Auria, J.C.; Tholl, D.; Ross, J.R.; Gershenzon, J.; Noel, J.P.; Pichersky, E. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 2003, 36, 577–588. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.; Zhou, Y.; Yu, Y.; Waseem, M.; Ashraf, U.; Li, X.; Yu, R.; Fan, Y. Genome-wide analysis of ARF transcription factors reveals HcARF5 expression profile associated with the biosynthesis of beta-ocimene synthase in Hedychium coronarium. Plant Cell Rep. 2021, 40, 1269–1284. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DELTADELTACT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Name | ID | Amino Acids Length | Molecular Weight (Da) | pI | Subfamily |
|---|---|---|---|---|---|
| HcBSMT1/HcSABATH1 | novel01829 | 382 | 43,017.53 | 6.11 | Clade VII |
| HcBSMT2/HcSABATH2 | scaf_368.32 | 377 | 43,032.38 | 5.46 | Clade VII |
| HcSABATH3 | scaf_68.69 | 378 | 42,554.68 | 5.65 | Clade VII |
| HcSABATH4 | scaf_368.54 | 377 | 41,940.34 | 6.04 | Clade VII |
| HcSABATH5 | scaf_368.25 | 379 | 38,197.02 | 5.40 | Clade VII |
| HcSABATH6 | scaf_102.154.1 | 376 | 42,220.80 | 5.06 | Clade VII |
| HcSABATH7 | scaf_102.154.23 | 378 | 42,919.37 | 5.63 | Clade VII |
| HcSABATH8 | scaf_102.155.1 | 373 | 42,418.53 | 5.70 | Clade VII |
| HcSABATH9 | scaf_102.155.2 | 366 | 41,399.01 | 5.08 | Clade VII |
| HcSABATH10 | scaf_462.93 | 387 | 43,220.25 | 4.96 | Clade V |
| HcJMT1/HcSABATH11 | scaf_72.107 | 376 | 41,686.44 | 5.42 | Clade V |
| HcJMT2/HcSABATH12 | novel01692 | 376 | 41,583.13 | 5.19 | Clade V |
| Substrates | Enzyme Activity * |
|---|---|
| Jasmonic acid | + |
| Dihydrojasmonic acid | + |
| Salicylic acid | − |
| Benzoic acid | − |
| 3-Hydroxybenzoic acid | − |
| 2,3-Dihydnoxybenzoic acid | − |
| Cinnamic acid | − |
| o-Anisic acid | − |
| o-Coumaric acid | − |
| Nicotinic acid | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, Y.; Zhang, X.; Wang, L.; He, J.; Yang, S.; Li, X.; Yu, Y.; Yu, R.; Fan, Y. Identification and Characterization of Jasmonic Acid Methyltransferase Involved in the Formation of Floral Methyl Jasmonate in Hedychium coronarium. Plants 2024, 13, 8. https://doi.org/10.3390/plants13010008
Yue Y, Zhang X, Wang L, He J, Yang S, Li X, Yu Y, Yu R, Fan Y. Identification and Characterization of Jasmonic Acid Methyltransferase Involved in the Formation of Floral Methyl Jasmonate in Hedychium coronarium. Plants. 2024; 13(1):8. https://doi.org/10.3390/plants13010008
Chicago/Turabian StyleYue, Yuechong, Xiaohong Zhang, Lan Wang, Jieling He, Shengnan Yang, Xinyue Li, Yunyi Yu, Rangcai Yu, and Yanping Fan. 2024. "Identification and Characterization of Jasmonic Acid Methyltransferase Involved in the Formation of Floral Methyl Jasmonate in Hedychium coronarium" Plants 13, no. 1: 8. https://doi.org/10.3390/plants13010008





