Genome-Wide Identification and Characterization of the TIFY Gene Family and Their Expression Patterns in Response to MeJA and Aluminum Stress in Centipedegrass (Eremochloa ophiuroides)
Abstract
:1. Introduction
2. Results
2.1. Identification of TIFY Family Genes in the E. ophiuroides Genome
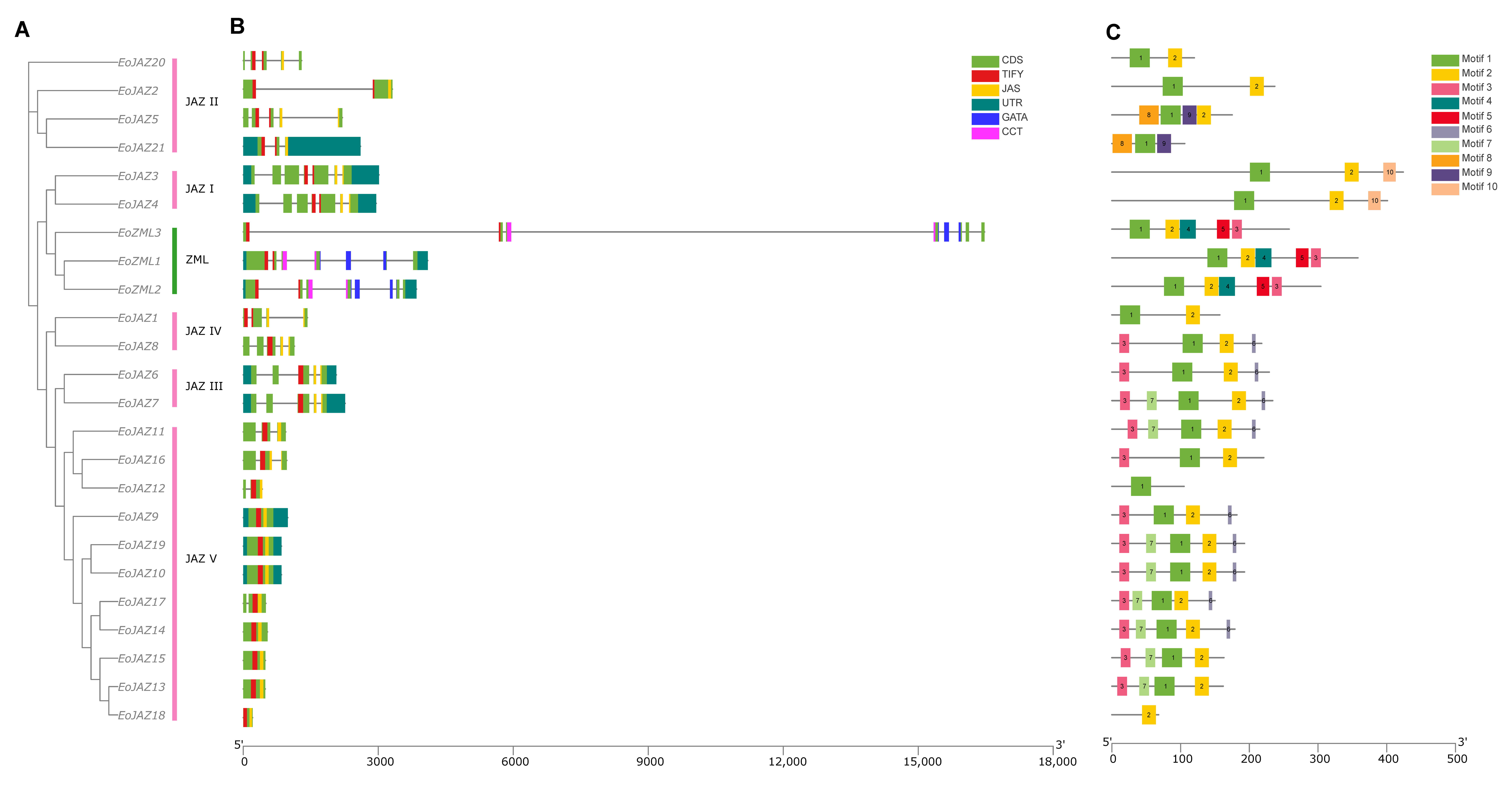
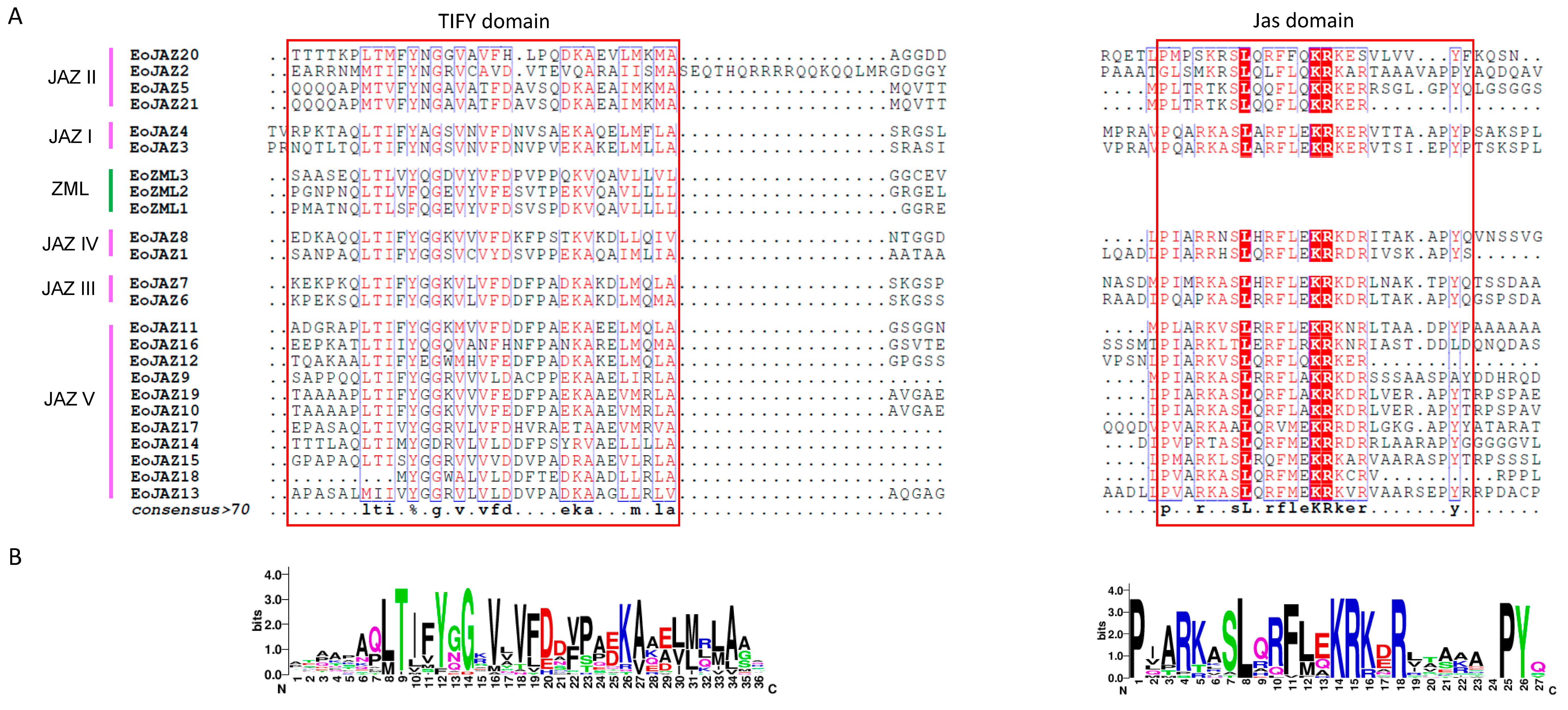
2.2. Multiple Sequence Alignment, Phylogenetic Analysis, and Classification of EoTIFY Proteins
2.3. Gene Structures and Conserved Motifs of EoTIFY Gene Family
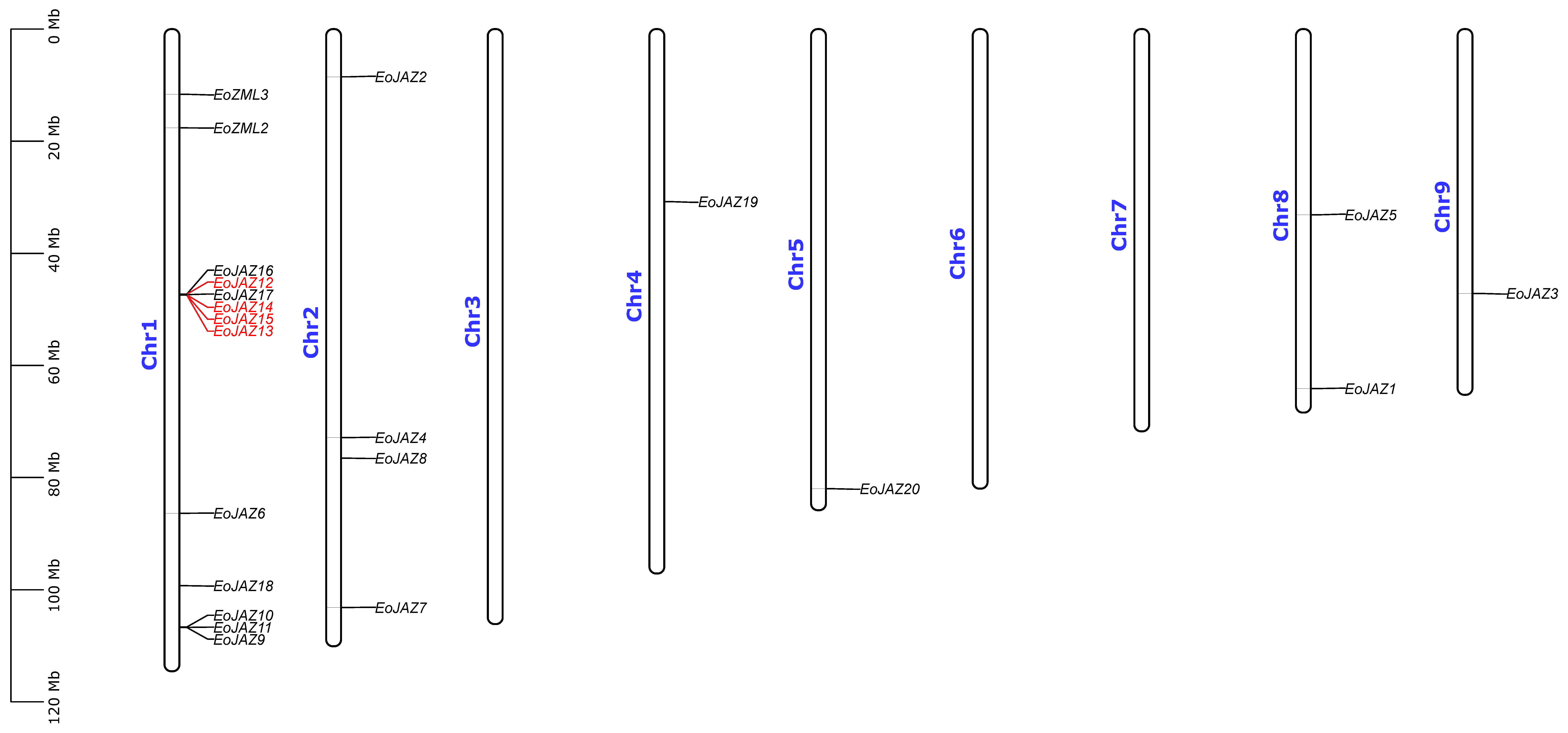
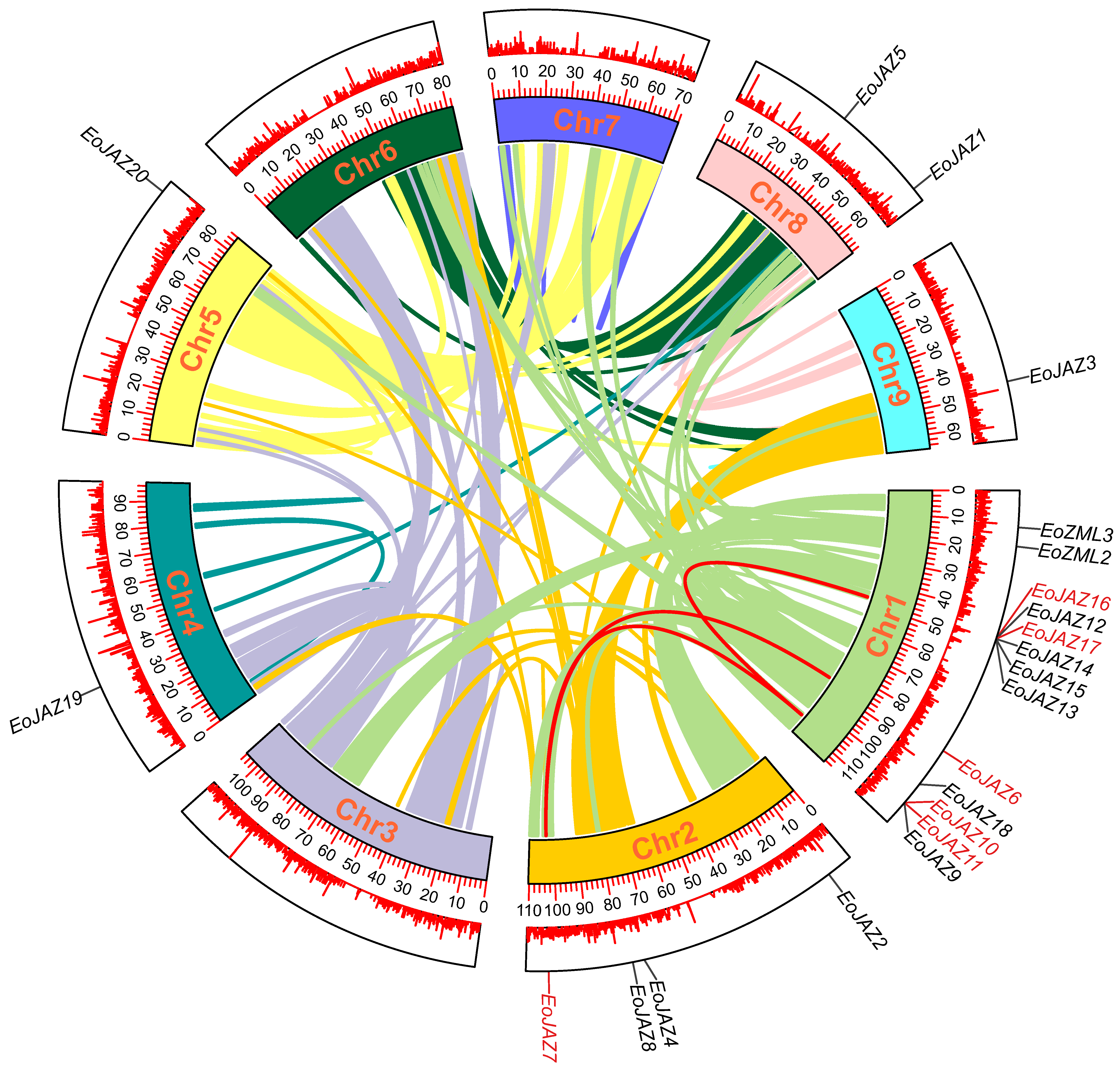
2.4. Chromosomal Distribution and Gene Duplication Analyses of the EoTIFY Genes
2.5. Synteny and Evolutionary Analyses of the E. ophiuroides TIFY Genes and Other Plants TIFYs
2.6. Expression Patterns of TIFY Family Genes in Various Tissues of E. ophiuroides
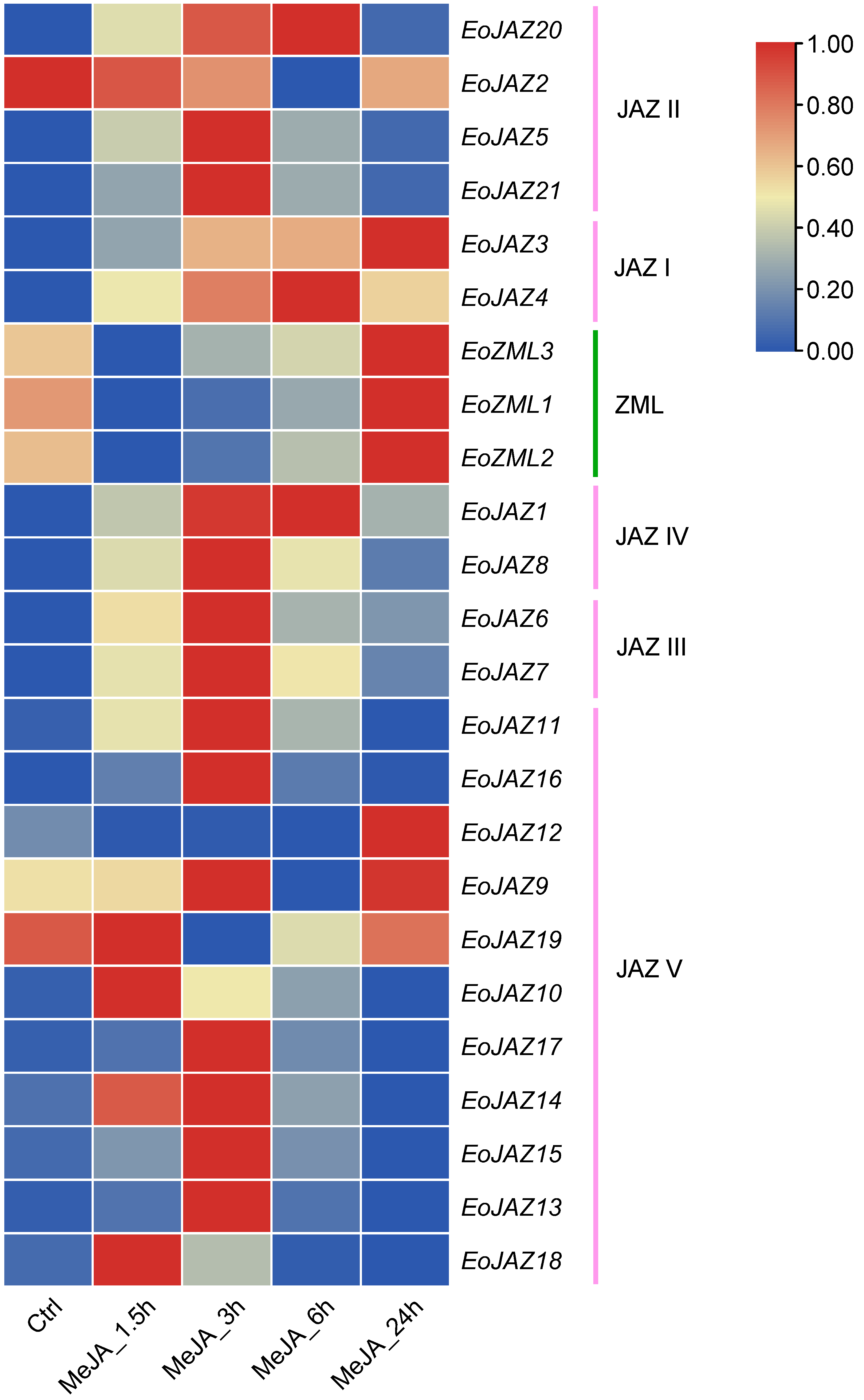
2.7. Expression Patterns of the EoTIFY Family Genes under MeJA Treatment
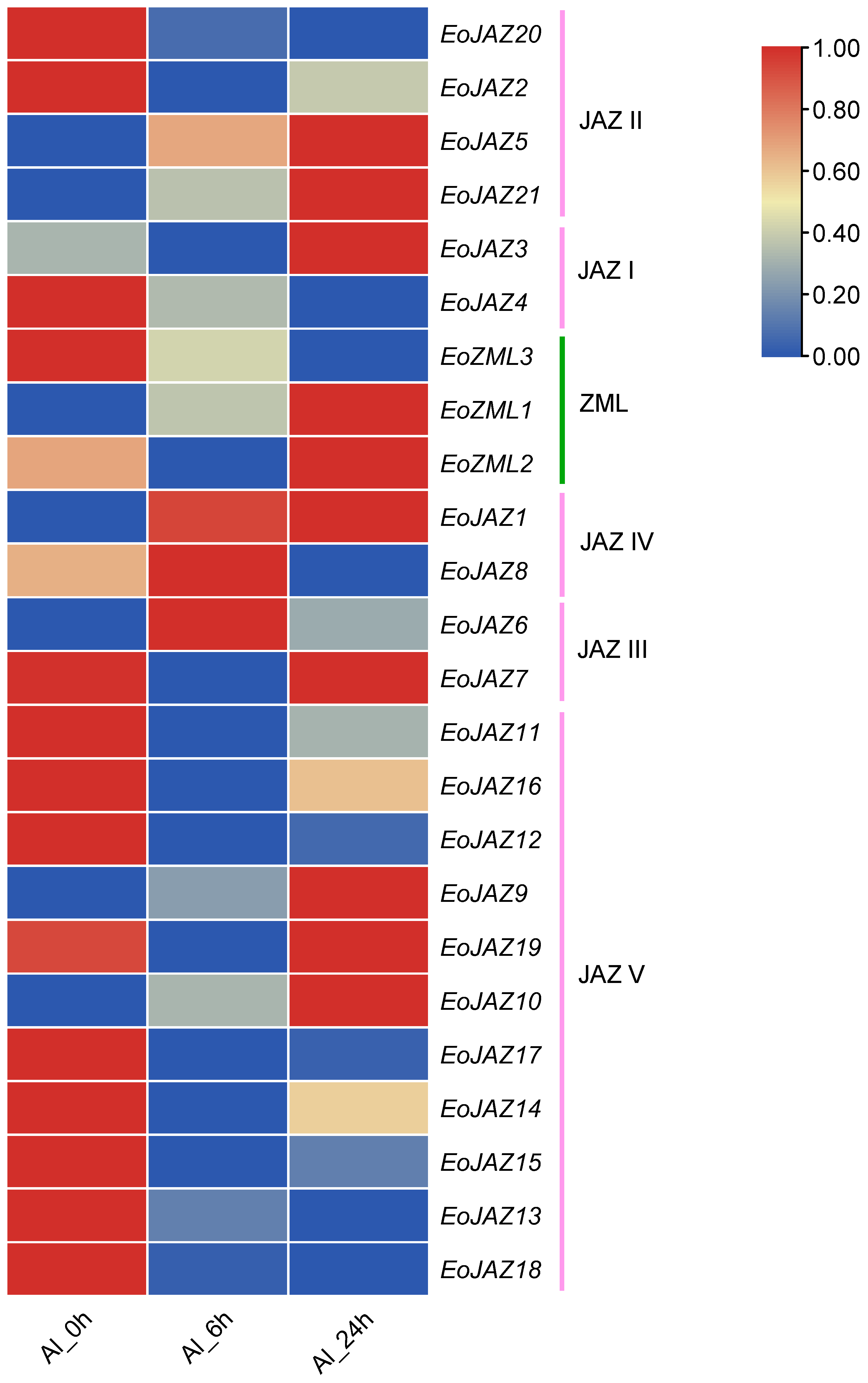
2.8. Expression Patterns of the EoTIFY Family Genes in Response to Aluminum Toxicity
3. Discussion
4. Materials and Methods
4.1. Identification of TIFY Gene Family Members in the E. ophiuroides Genome
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Gene Structure Analysis and Conserved Motif Discovery
4.4. Chromosomal Locations, Gene Duplication, and Synteny Analysis
4.5. Plant Materials and Stress Treatments
4.6. RNA Isolation and cDNA Synthesis
4.7. Primer Design and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Islam, M.; Hirata, M. Centipedegrass (Eremochloa ophiuroides (Munro) Hack.): Growth behavior and multipurpose usages. Grassl. Sci. 2005, 51, 183–190. [Google Scholar] [CrossRef]
- Li, J.; Guo, H.; Zong, J.; Chen, J.; Li, D.; Liu, J. Genetic diversity in centipedegrass [Eremochloa ophiuroides (Munro) Hack.]. Hortic. Res. 2020, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, A.; Wang, T.; Wang, Y.; Li, L.; Wu, P. Characterization and coexpression analysis of the Tify family genes in euryale ferox related to leaf development. Plants 2023, 12, 2323. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The TIFY family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E. JAZing up jasmonate signaling. Trends Plant Sci. 2008, 13, 66–71. [Google Scholar] [CrossRef]
- Chung, H.S.; Niu, Y.J.; Browse, J.; Howe, G.A. Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 2009, 70, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, F.; Chen, D.; Chu, W.; Liu, H.; Xiang, Y. Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiol. Biochem. 2017, 115, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Matsuda, Y.; Ando, K.; Nishii, A.; Takemura, M.; Yokota, A.; Kohchi, T. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004, 55, 631–639. [Google Scholar] [CrossRef]
- White, D.W. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 13238–13243. [Google Scholar] [CrossRef]
- Hakata, M.; Kuroda, M.; Ohsumi, A.; Hirose, T.; Nakamura, H.; Muramatsu, M.; Ichikawa, H. Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 2012, 76, 2129–2134. [Google Scholar] [CrossRef]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef] [PubMed]
- Demianski, A.J.; Chung, K.M.; Kunkel, B.N. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant. Pathol. 2012, 13, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, S.; Hosokawa-Shinonaga, Y.; Tamaoki, D.; Yamada, S.; Akimitsu, K.; Gomi, K. Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 2014, 37, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Liu, J.; Liu, P.; Ming, D.; Sun, J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 2019, 9, 5691. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Chen, L.; Li, J.; Hu, M.; Ullah, A.; He, X.; Yang, X.; Zhang, X. The JASMONATE ZIM-domain gene family mediates JA signaling and stress response in cotton. Plant Cell Physiol. 2017, 58, 2139–2154. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Li, R.; Liu, X.; Sun, M.; Wu, J.; Zhang, N.; Zhu, Y. The positive regulatory roles of the TIFY10 proteins in plant responses to alkaline stress. PLoS ONE 2014, 9, e111984. [Google Scholar] [CrossRef] [PubMed]
- Peethambaran, P.K.; Glenz, R.; Höninger, S.; Islam, S.M.S.; Hummel, S.; Harter, K.; Kolukisaoglu, U.; Meynard, D.; Guiderdoni, E.; Nick, P.; et al. Salt-inducible expression of OsJAZ8 improves resilience against salt-stress. BMC Plant Biol. 2018, 18, 311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, M.; Singer, S.D.; Fei, Z.; Wang, H.; Wang, X. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 2012, 7, e44465. [Google Scholar] [CrossRef]
- Sirhindi, G.; Sharma, P.; Arya, P.; Goel, P.; Kumar, G.; Acharya, V.; Singh, A.K. Genome-wide characterization and expression profiling of TIFY gene family in pigeonpea (Cajanus cajan (L.) Millsp.) under copper stress. J. Plant Biochem. Biotechnol. 2016, 25, 301–310. [Google Scholar] [CrossRef]
- Xiao, D.; Li, X.; Zhou, Y.Y.; Wei, L.; Keovongkod, C.; He, H.; Zhan, J.; Wang, A.Q.; He, L.F. Transcriptome analysis reveals significant difference in gene expression and pathways between two peanut cultivars under Al stress. Gene 2021, 781, 145535. [Google Scholar] [CrossRef]
- Xie, S.; Cui, L.; Lei, X.; Yang, G.; Li, J.; Nie, X.; Ji, W. The TIFY gene family in wheat and its progenitors: Genome-wide identification, evolution and expression analysis. Curr. Genom. 2019, 20, 371–388. [Google Scholar] [CrossRef]
- Singh, P.; Mukhopadhyay, K. Comprehensive molecular dissection of TIFY transcription factors reveal their dynamic responses to biotic and abiotic stress in wheat (Triticum aestivum L.). Sci. Rep. 2021, 11, 9739. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Yu, R.; Han, M.; Wu, Z. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family. Mol. Genet. Genom. 2015, 290, 1849–1858. [Google Scholar] [CrossRef] [PubMed]
- Heidari, P.; Faraji, S.; Ahmadizadeh, M.; Ahmar, S.; Mora-Poblete, F. New insights into structure and function of TIFY genes in Zea mays and Solanum lycopersicum: A genome-wide comprehensive analysis. Front. Genet. 2021, 12, 657970. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Fang, Y.; Jiang, J.; Chen, M.; Li, X.; Xin, X. Genome-wide identification and characterization of the JAZ gene family and its expression patterns under various abiotic stresses in Sorghum bicolor. J. Integr. Agric. 2022, 21, 3540–3555. [Google Scholar] [CrossRef]
- Shrestha, K.; Huang, Y. Genome-wide characterization of the sorghum JAZ gene family and their responses to phytohormone treatments and aphid infestation. Sci. Rep. 2022, 12, 3238. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zi, H.; Wang, R.; Liu, J.; Wang, H.; Chen, R.; Li, L.; Guo, H.; Chen, J.; Li, J.; et al. A high-quality chromosome-scale assembly of the centipedegrass [Eremochloa ophiuroides (Munro) Hack.] genome provides insights into chromosomal structural evolution and prostrate growth habit. Hortic. Res. 2021, 8, 201. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Ahmad, M.; Zahir, Z.A.; Kharal, M.A. Role of phytohormones in stress tolerance of plants. In Plant, Soil and Microbes; Springer: Berlin/Heidelberg, Germany, 2016; pp. 385–421. [Google Scholar]
- Thireault, C.; Shyu, C.; Yoshida, Y.; Aubin, B.S.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef]
- Lv, G.; Han, R.; Shi, J.; Chen, K.; Liu, G.; Yu, Q.; Yang, C.; Jiang, J. Genome-wide identification of the TIFY family reveals JAZ subfamily function in response to hormone treatment in Betula platyphylla. BMC Plant Biol. 2023, 23, 143. [Google Scholar] [CrossRef]
- Monte, I.; Franco-Zorrilla, J.M.; García-Casado, G.; Zamarreno, A.M.; García-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano1, R. A single JAZ repressor controls the Jasmonate pathway in Marchantia polymorpha. Mol. Plant 2019, 12, 185–198. [Google Scholar] [CrossRef]
- Zhang, X.; Ran, W.; Zhang, J.; Ye, M.; Lin, S.; Li, X.; Sultana, R.; Sun, X. Genome-wide identification of the Tify gene family and their expression profiles in response to biotic and abiotic stresses in tea plants (Camellia sinensis). Int. J. Mol. Sci. 2020, 21, 8316. [Google Scholar] [CrossRef]
- Zhang, L.; You, J.; Chan, Z. Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 2015, 128, 995–1005. [Google Scholar] [CrossRef]
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef]
- Huang, Z.; Jin, S.H.; Guo, H.D.; Zhong, X.J.; He, J.; Li, X.; Jiang, M.; Yu, X.; Long, H.; Ma, M.; et al. Genome-wide identification and characterization of TIFY family genes in Moso Bamboo (Phyllostachys edulis) and expression profiling analysis under dehydration and cold stresses. PeerJ 2016, 4, e2620. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Jia, H.; Wu, M.; Zhong, W.; Jia, D.; Wang, Z.; Huang, C. Genome-wide identification and characterization of the TIFY gene family in kiwifruit. BMC Genom. 2022, 23, 179. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Grigoriev, A. Protein domains correlate strongly with exons in multiple eukaryotic genomes–evidence of exon shuffling? Trends Genet. 2004, 20, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xiang, Y.; Fang, L.; Wang, Y.; Xin, H.; Li, S. Patterns of gene duplication and their contribution to expansion of gene families in grapevine. Plant Mol. Biol. Rep. 2013, 31, 852–861. [Google Scholar] [CrossRef]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Kim, K.C.; Han, J.A.; Lee, J.; Maeng, J.; Hur, Y. Gene encoding PnFL-2 with TIFY and CCT motifs may control floral induction in Pharbitis nil. Genes Genom. 2011, 33, 229–236. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; Riemann, M.; Nick, P. The jasmonate pathway mediates salt tolerance in grapevines. J. Exp. Bot. 2012, 63, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Overexpression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Ogawa, T.; Hashimoto, T. Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol. 2008, 49, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Q.; Jiang, H.L.; Li, C.Y. Systemin/jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 2011, 4, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Baldwin, I.T.; Gális, I. NaJAZh regulates a subset of defense responses against herbivores and spontaneous leaf necrosis in Nicotiana attenuata plants. Plant Physiol. 2012, 159, 769–788. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Koo, A.J.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef]
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Chen, Y.; He, Q.; Li, X.; Zhang, Y.; Li, J.; Zhang, L.; Yao, X.; Zhang, X.; Liu, C.; Wang, H. Identification of accurate reference genes for qRT-PCR analysis of gene expression in Eremochloa ophiuroides under multiple stresses of phosphorus deficiency and/or aluminum toxicity. Plants 2023, 12, 3751. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nature Protocols 2008, 3, 1101. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zhang, Y.; Zhang, L.; Li, X.; Yao, X.; Hao, D.; Guo, H.; Liu, J.; Li, J. Genome-Wide Identification and Characterization of the TIFY Gene Family and Their Expression Patterns in Response to MeJA and Aluminum Stress in Centipedegrass (Eremochloa ophiuroides). Plants 2024, 13, 462. https://doi.org/10.3390/plants13030462
Wang H, Zhang Y, Zhang L, Li X, Yao X, Hao D, Guo H, Liu J, Li J. Genome-Wide Identification and Characterization of the TIFY Gene Family and Their Expression Patterns in Response to MeJA and Aluminum Stress in Centipedegrass (Eremochloa ophiuroides). Plants. 2024; 13(3):462. https://doi.org/10.3390/plants13030462
Chicago/Turabian StyleWang, Haoran, Yuan Zhang, Ling Zhang, Xiaohui Li, Xiang Yao, Dongli Hao, Hailin Guo, Jianxiu Liu, and Jianjian Li. 2024. "Genome-Wide Identification and Characterization of the TIFY Gene Family and Their Expression Patterns in Response to MeJA and Aluminum Stress in Centipedegrass (Eremochloa ophiuroides)" Plants 13, no. 3: 462. https://doi.org/10.3390/plants13030462




