Emerging Roles of Epigenetics in Grapevine and Winegrowing
Abstract
:1. Introduction
2. Overview of Epigenetic Mechanisms in Plants
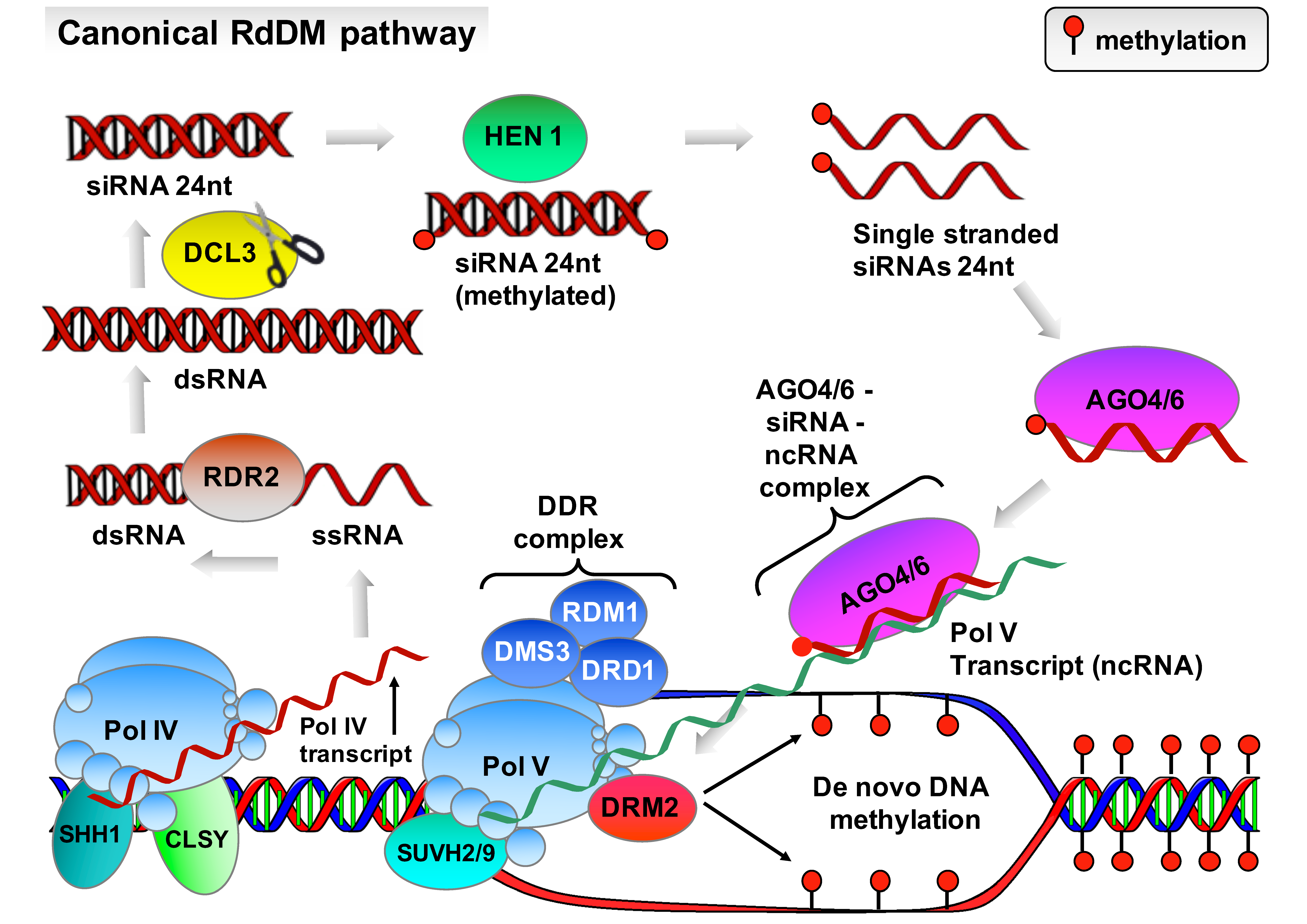
2.1. DNA Methylation
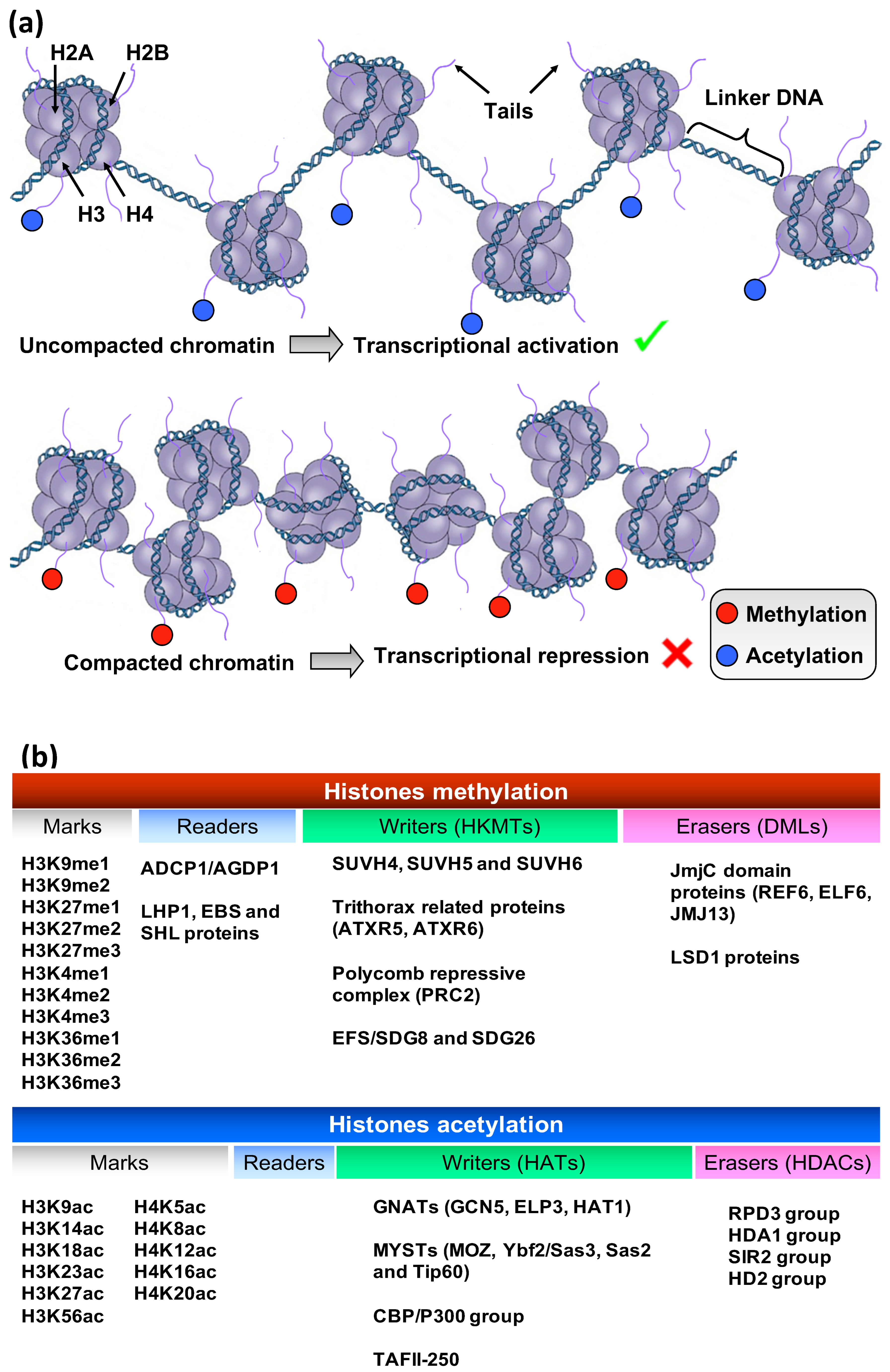
2.2. Histones Post-Translational Modifications (HPTMs)
3. Epigenetic Changes Due to Adverse Environmental Conditions
4. Epigenetic Effects on Berry Development and Quality
5. Effects of Grafting on Grapevine Epigenome
6. Epigenetics and Defense Responses to Pathogens
7. Epigenetic Memory
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Alston, J.M.; Sambucci, O. Grapes in the world economy. In The Grape Genome; Cantu, D., Walker, M.A., Eds.; Springer: New Delhi, India, 2019; pp. 1–24. [Google Scholar]
- Banilas, G.; Korkas, E.; Kaldis, P.; Hatzopoulos, P. Olive and grapevine biodiversity in Greece and Cyprus-A review. In Sustainable Agriculture Reviews; Lichtfouse, E., Ed.; Springer: Dordrecht, The Netherlands, 2009; Volume 2, pp. 401–428. [Google Scholar]
- Armijo, G.; Schlechter, R.; Agurto, M.; Muñoz, D.; Nuñez, C.; Arce-Johnson, P. Grapevine pathogenic microorganisms: Understanding infection strategies and host response scenarios. Front. Plant Sci. 2016, 7, 382. [Google Scholar] [CrossRef]
- Gambetta, G.A.; Herrera, J.C.; Dayer, S.; Feng, Q.; Hochberg, U.; Castellarin, S.D. The physiology of drought stress in grapevine: Towards an integrative definition of drought tolerance. J. Exp. Bot. 2020, 71, 4658–4676. [Google Scholar] [CrossRef]
- Venios, X.; Korkas, E.; Nisiotou, A.; Banilas, G. Grapevine responses to heat stress and global warming. Plants 2020, 9, 1754. [Google Scholar] [CrossRef]
- Santos, R.B.; Figueiredo, A. Two sides of the same story in grapevine–pathogen interactions. J. Exp. Bot. 2021, 72, 3367–3380. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Evolutionary significance of phenotypic plasticity in plants. Adv. Genet. 1965, 13, 115–155. [Google Scholar]
- Pigliucci, M. Evolution of phenotypic plasticity: Where are we going now? Trends Ecol. Evol. 2005, 20, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Dar, F.A.; Mushtaq, N.U.; Saleem, S.; Rehman, R.U.; Dar, T.U.H.; Hakeem, K.R. Role of epigenetics in modulating phenotypic plasticity against abiotic stresses in plants. Int. J. Genom. 2022, 2022, 1092894. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.E.; Nota, F.; Cambiagno, D.A. Epigenetic control of plant immunity. Mol. Plant Pathol. 2010, 11, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Kovalchuk, I. Epigenetic control of plant stress response. Environ. Mol. Mutagen. 2008, 49, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Espinas, N.A.; Saze, H.; Saijo, Y. Epigenetic control of defense signaling and priming in plants. Front. Plant Sci. 2016, 7, 1201. [Google Scholar] [CrossRef] [PubMed]
- Gkizi, D.; González Gil, A.; Pardal, A.J.; Piquerez, S.J.; Sergaki, C.; Ntoukakis, V.; Tjamos, S.E. The bacterial biocontrol agent Paenibacillus alvei K165 confers inherited resistance to Verticillium dahliae. J. Exp. Bot. 2021, 72, 4565–4576. [Google Scholar] [CrossRef]
- Harris, Z.N.; Pratt, J.E.; Kovacs, L.G.; Klein, L.L.; Kwasniewski, M.T.; Londo, J.P.; Wu, A.S.; Miller, A.J. Grapevine scion gene expression is driven by rootstock and environment interaction. BMC Plant Biol. 2023, 23, 211. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.M.; Stammitti, L.; Carrillo, N.; Blancquaert, E.; Rubio, B.; Teyssier, E.; Gallusci, P. Epigenetics: An innovative lever for grapevine breeding in times of climatic changes. OENO One 2023, 57, 265–282. [Google Scholar] [CrossRef]
- Vaucheret, H.; Fagard, M. Transcriptional gene silencing in plants: Targets, inducers and regulators. Trends Genet. 2001, 17, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Sijen, T.; Vijn, I.; Rebocho, A.; van Blokland, R.; Roelofs, D.; Mol, J.N.; Kooter, J.M. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 2001, 11, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolomé, J. DNA methylation in plants: Mechanisms and tools for targeted manipulation. New Phytol. 2020, 227, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Hannan Parker, A.; Wilkinson, S.W.; Ton, J. Epigenetics: A catalyst of plant immunity against pathogens. New Phytol. 2022, 233, 66–83. [Google Scholar] [CrossRef]
- Liu, J.; He, Z. Small DNA methylation, big player in plant abiotic stress responses and memory. Front. Plant Sci. 2020, 11, 595603. [Google Scholar] [CrossRef]
- Mahfouz, M.M. RNA-directed DNA methylation: Mechanisms and functions. Plant Signal. Behav. 2010, 5, 806–816. [Google Scholar] [CrossRef]
- Tirnaz, S.; Batley, J. Epigenetics: Potentials and challenges in crop breeding. Mol. Plant. 2019, 12, 1309–1311. [Google Scholar] [CrossRef] [PubMed]
- Lizamore, D.; Bicknell, R.; Winefield, C. Elevated transcription of transposable elements is accompanied by het-siRNA-driven de novo DNA methylation in grapevine embryogenic callus. BMC Genom. 2021, 22, 676. [Google Scholar] [CrossRef]
- Pereira, G.; Pereira, J.; Santos, R.B.; Figueiredo, A. Uncovering the role of DNA methyltransferases in grapevine—Plasmopara viticola interaction: From genome-wide characterization to global methylation patterns. Gene 2022, 837, 146693. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Moritoh, S.; Johzuka-Hisatomi, Y.; Ono, A.; Terada, R.; Nakamura, I.; Iida, S. Alternative splicing of the rice OsMET1 genes encoding maintenance DNA methyltransferase. J. Plant Physiol. 2008, 165, 1774–1782. [Google Scholar] [CrossRef]
- Zhang, X.; Jacobsen, S.E. Genetic analyses of DNA methyltransferases in Arabidopsis thaliana. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2006; Volume 71, pp. 439–447. [Google Scholar]
- Ahmad, F.; Huang, X.; Lan, H.X.; Huma, T.; Bao, Y.M.; Huang, J.; Zhang, H.S. Comprehensive gene expression analysis of the DNA (cytosine-5) methyltransferase family in rice (Oryza sativa L.). Genet. Mol. Res. 2014, 13, 5159–5172. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Zhou, M.M. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell. Mol. Life Sci. 2006, 63, 2755–2763. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA methylation. PLoS Genet. 2020, 16, 1009034. [Google Scholar] [CrossRef]
- Blevins, T.; Podicheti, R.; Mishra, V.; Marasco, M.; Wang, J.; Rusch, D.; Tang, H.; Pikaard, C.S. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 2015, 4, 9591. [Google Scholar] [CrossRef]
- Liu, W.; Shoji, K.; Naganuma, M.; Tomari, Y.; Iwakawa, H.O. The mechanisms of siRNA selection by plant Argonaute proteins triggering DNA methylation. Nucleic Acids Res. 2022, 50, 12997–13010. [Google Scholar] [CrossRef]
- Xie, M.; Yu, B. siRNA-directed DNA methylation in plants. Curr. Genomics. 2015, 16, 23–31. [Google Scholar] [CrossRef]
- Wongpalee, S.P.; Liu, S.; Gallego-Bartolomé, J.; Leitner, A.; Aebersold, R.; Liu, W.; Yen, L.; Nohales, M.A.; Kuo, P.H.; Vashisht, A.A.; et al. CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation. Nat. Commun. 2019, 10, 3916. [Google Scholar] [CrossRef] [PubMed]
- McCue, A.D.; Panda, K.; Nuthikattu, S.; Choudury, S.G.; Thomas, E.N.; Slotkin, R.K. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015, 34, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Shen, R.; Liu, X.; Yang, X.; Xie, K.; Guo, Z.; Wang, D. Histone post-translational modification and the DNA damage response. Genes Dis. 2022, 10, 1429–1444. [Google Scholar] [CrossRef]
- Fortes, A.M.; Gallusci, P. Plant stress responses and phenotypic plasticity in the epigenomics era: Perspectives on the grapevine scenario, a model for perennial crop plants. Front. Plant Sci. 2017, 8, 82. [Google Scholar] [CrossRef]
- Zhao, T.; Zhan, Z.; Jiang, D. Histone modifications and their regulatory roles in plant development and environmental memory. J. Genet. Genom. 2019, 46, 467–476. [Google Scholar] [CrossRef]
- Aquea, F.; Timmermann, T.; Arce-Johnson, P. Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol. Biochem. 2010, 48, 194–199. [Google Scholar] [CrossRef]
- Qi, Q.; Hu, B.; Jiang, W.; Wang, Y.; Yan, J.; Ma, F.; Guan, Q.; Xu, J. Advances in plant epigenome editing research and its application in plants. Int. J. Mol. Sci. 2023, 24, 3442. [Google Scholar] [CrossRef]
- Gujral, P.; Mahajan, V.; Lissaman, A.C.; Ponnampalam, A.P. Histone acetylation and the role of histone deacetylases in normal cyclic endometrium. Reprod. Biol. Endocrinol. 2020, 18, 84. [Google Scholar] [CrossRef]
- Williams, B.R.; Edwards, C.E.; Kwasniewski, M.T.; Miller, A.J. Epigenomic patterns reflect irrigation and grafting in the grapevine clone ‘Chambourcin’. arXiv 2020. [Google Scholar] [CrossRef]
- Fabres, P.J. A Multiple ‘Omics’ Approach to Study the Interaction between the Vitis vinifera Transcriptome and Epigenome and the Barossa Valley Terroir. Ph.D. Thesis, The University of Adelaide, Adelaide, Australia, 2020. [Google Scholar]
- Tan, J.W.; Shinde, H.; Tesfamicael, K.; Hu, Y.; Fruzangohar, M.; Tricker, P.; Baumann, U.; Edwards, E.J.; Rodríguez López, C.M. Global transcriptome and gene co-expression network analyses reveal regulatory and non-additive effects of drought and heat stress in grapevine. Front. Plant Sci. 2023, 14, 1096225. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, Q.; Gichuki, D.K.; Hou, Y.; Liu, Y.; Zhou, H.; Xu, C.; Fang, L.; Gong, L.; Zheng, B.; et al. Genome-wide profiling of histone H3 lysine 27 trimethylation and its modification in response to chilling stress in grapevine leaves. Hortic. Plant J. 2023, 9, 496–508. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Z.; Sadeghnezhad, E.; Pang, Q.; Li, S.; Pervaiz, T.; Su, Z.; Dong, T.; Fang, J.; Jia, H. Demethylation alters transcriptome profiling of buds and leaves in ‘Kyoho’ grape. BMC Plant Biol. 2020, 20, 544. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, V.; Falchi, R.; Peressotti, A.; Vizzotto, G. Expression patterns of DEMETER-like DNA demethylases homologs hint at potential involvement in grapevine dormancy release. BIO Web Conf. 2022, 44, 4001. [Google Scholar] [CrossRef]
- Sun, X.; Fan, G.; Su, L.; Wang, W.; Liang, Z.; Li, S.; Xin, H. Identification of cold-inducible microRNAs in grapevine. Front. Plant Sci. 2015, 6, 595. [Google Scholar] [CrossRef] [PubMed]
- Varela, A.; Ibañez, V.N.; Alonso, R.; Zavallo, D.; Asurmendi, S.; Gomez Talquenca, S.; Marfil, C.F.; Berli, F.J. Vineyard environments influence Malbec grapevine phenotypic traits and DNA methylation patterns in a clone-dependent way. Plant Cell Rep. 2021, 40, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Baránková, K.; Nebish, A.; Tříska, J.; Raddová, J.; Baránek, M. Comparison of DNA methylation landscape between Czech and Armenian vineyards show their unique character and increased diversity. Czech J. Genet. Plant Breed. 2021, 57, 67–75. [Google Scholar] [CrossRef]
- Xie, H.; Konate, M.; Sai, N.; Tesfamicael, K.G.; Cavagnaro, T.; Gilliham, M.; Breen, J.; Metcalfe, A.; Stephen, J.R.; De Bei, R.; et al. Global DNA methylation patterns can play a role in defining terroir in grapevine (Vitis vinifera cv. Shiraz). Front. Plant Sci. 2017, 8, 1860. [Google Scholar] [CrossRef]
- Tan, J.; Lopez, C.M.R. Epigenomics: A new tool for the generation of climate resilient grapevines. Front. Hortic. 2023, 2, 1116866. [Google Scholar] [CrossRef]
- Fraga, M.F.; Rodríguez, R.; Cañal, M.J. Genomic DNA methylation–demethylation during aging and reinvigoration of Pinus radiata. Tree Physiol. 2002, 22, 813–816. [Google Scholar] [CrossRef]
- Valledor, L.; Hasbún, R.; Meijón, M.; Rodríguez, J.L.; Santamaría, E.; Viejo, M.; Berdasco, M.; Feito, I.; Fraga, M.F.; Canal, M.J.; et al. Involvement of DNA methylation in tree development and micropropagation. Plant Cell Tissue Organ Cult. 2007, 91, 75–86. [Google Scholar] [CrossRef]
- Baránek, M.; Čechová, J.; Raddová, J.; Holleinová, V.; Ondrušíková, E.; Pidra, M. Dynamics and reversibility of the DNA methylation landscape of grapevine plants (Vitis vinifera) stressed by in vitro cultivation and thermotherapy. PLoS ONE 2015, 10, e0126638. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Kiselev, K.V. Age-associated alterations in the somatic mutation and DNA methylation levels in plants. Plant Biol. 2016, 18, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Marfil, C.; Ibañez, V.; Alonso, R.; Varela, A.; Bottini, R.; Masuelli, R.; Fontana, A.; Berli, F. Changes in grapevine DNA methylation and polyphenols content induced by solar ultraviolet-B radiation, water deficit and abscisic acid spray treatments. Plant Physiol. Biochem. 2019, 135, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, V.; Vitali, M.; Boccacci, P.; Miozzi, L.; Cuozzo, D.; Chitarra, W.; Mannini, F.; Lovisolo, C.; Gambino, G. Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Sci. Rep. 2016, 6, 20167. [Google Scholar] [CrossRef]
- Xie, H.; Konate, M.; Sai, N.; Tesfamicael, K.G.; Cavagnaro, T.; Gilliham, M.; Breen, J.; Metcalfe, A.; DeBei, R.; Collins, C.; et al. Environmental conditions and agronomic practices induce consistent global changes in DNA methylation patterns in grapevine (Vitis vinifera cv. Shiraz). arXiv 2017, arXiv:127977. [Google Scholar]
- Berli, F.J.; Moreno, D.; Piccoli, P.; Hespanhol-Viana, L.; Silva, M.F.; Bressan-Smith, R.; Cavagnaro, J.B.; Bottini, R. Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet-B radiation by enhancing ultraviolet-absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ. 2010, 33, 1–10. [Google Scholar]
- Carbonell-Bejerano, P.; Diago, M.P.; Martínez-Abaigar, J.; Martínez-Zapater, J.M.; Tardáguila, J.; Núñez-Olivera, E. Solar ultraviolet radiation is necessary to enhance grapevine fruit ripening transcriptional and phenolic responses. BMC Plant Biol. 2014, 14, 183. [Google Scholar] [CrossRef]
- Tyunin, A.P.; Kiselev, K.V. Alternations in VaSTS gene cytosine methylation and t-resveratrol production in response to UV-C irradiation in Vitis amurensis Rupr. cells. Plant Cell Tissue Organ Cult. 2016, 124, 33–45. [Google Scholar] [CrossRef]
- Dal Santo, S.; Tornielli, G.B.; Zenoni, S.; Fasoli, M.; Farina, L.; Anesi, A.; Guzzo, F.; Delledonne, M.; Pezzotti, M. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013, 14, r54. [Google Scholar] [CrossRef]
- Xu, F.; Xi, Z.M.; Zhang, H.; Zhang, C.J.; Zhang, Z.W. Brassinosteroids are involved in controlling sugar unloading in Vitis vinifera ‘Cabernet Sauvignon’ berries during véraison. Plant Physiol. Biochem. 2015, 94, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Babalık, Z.; Demirci, T.; Aşcı, Ö.A.; Baydar, N.G. Brassinosteroids modify yield, quality, and antioxidant components in grapes (Vitis vinifera cv. Alphonse Lavallée). J. Plant Growth Regul. 2020, 39, 147–156. [Google Scholar] [CrossRef]
- Kong, Y.; Olejar, K.J.; On, S.L.; Winefield, C.; Wescombe, P.A.; Brennan, C.S.; Hider, R.N.; Chelikani, V. Epigenetic changes in Saccharomyces cerevisiae alters the aromatic profile in alcoholic fermentation. Appl. Environ. Microbiol. 2022, 88, e01528-22. [Google Scholar] [CrossRef] [PubMed]
- Aquea, F.; Vega, A.; Timmermann, T.; Poupin, M.J.; Arce-Johnson, P. Genome-wide analysis of the SET DOMAIN GROUP family in grapevine. Plant Cell Rep. 2011, 30, 1087–1097. [Google Scholar] [CrossRef]
- Lü, P.; Yu, S.; Zhu, N.; Chen, Y.R.; Zhou, B.; Pan, Y.; Tzeng, D.; Fabi, J.P.; Argyris, J.; Garcia-Mas, J.; et al. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants 2018, 4, 784–791. [Google Scholar] [CrossRef]
- Tirumalai, V.; Swetha, C.; Nair, A.; Pandit, A.; Shivaprasad, P.V. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 2019, 70, 4775–4792. [Google Scholar] [CrossRef]
- Teixeira, A.; Eiras-Dias, J.; Castellarin, S.D.; Gerós, H. Berry phenolics of grapevine under challenging environments. Int. J. Mol. Sci. 2013, 14, 18711–18739. [Google Scholar] [CrossRef] [PubMed]
- Tyunin, A.P.; Kiselev, K.V.; Karetin, Y.A. Differences in the methylation patterns of the VaSTS1 and VaSTS10 genes of Vitis amurensis. Biotechnol. Lett. 2013, 35, 1525–1532. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Tyunin, A.P.; Zhuravlev, Y.N. Involvement of DNA methylation in the regulation of STS10 gene expression in Vitis amurensis. Planta 2013, 237, 933–941. [Google Scholar] [CrossRef]
- Xia, H.; Shen, Y.; Hu, R.; Wang, J.; Deng, H.; Lin, L.; Liang, D. Methylation of MYBA1 is associated with the coloration in ‘Manicure Finger’ grape skin. Agric. Food Chem. 2021, 69, 15649–15659. [Google Scholar] [CrossRef]
- Azuma, A.; Kobayashi, S. Demethylation of the 3′ LTR region of retrotransposon in VvMYBA1BEN allele enhances anthocyanin biosynthesis in berry skin and flesh in ‘Brazil’ grape. Plant Sci. 2022, 322, 111341. [Google Scholar] [CrossRef]
- Gomès, É.; Maillot, P.; Duchêne, É. Molecular tools for adapting viticulture to climate change. Front. Plant Sci. 2021, 12, 633846. [Google Scholar] [CrossRef] [PubMed]
- Powell, K.S. Grape phylloxera: An overview. In Root Feeders: An Ecosystem Approach; Johnson, S.N., Murray, P.J., Eds.; Integrative Approaches for the Investigation of Root Herbivory in Agricultural and Natural Systems: Berkshire, UK, 2004; pp. 96–114. [Google Scholar]
- Wang, J.; Jiang, L.; Wu, R. Plant grafting: How genetic exchange promotes vascular reconnection. New Phytol. 2017, 214, 56–65. [Google Scholar] [CrossRef]
- Anesi, A.; Stocchero, M.; Dal Santo, S.; Commisso, M.; Zenoni, S.; Ceoldo, S.; Guzzo, F. Towards a scientific interpretation of the terroir concept: Plasticity of the grape berry metabolome. BMC Plant Biol. 2015, 15, 191. [Google Scholar] [CrossRef] [PubMed]
- Migicovsky, Z.; Harris, Z.N.; Klein, L.L.; Li, M.; McDermaid, A.; Chitwood, D.H.; Fennell, A.; Kovacs, L.G.; Kwasniewski, M.; Londo, J.P.; et al. Rootstock effects on scion phenotypes in a ‘Chambourcin’ experimental vineyard. Hortic. Res. 2019, 6, 64. [Google Scholar] [CrossRef]
- Yang, Y.; Mao, L.; Jittayasothorn, Y.; Kang, Y.; Jiao, C.; Fei, Z.; Zhong, G.Y. Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol. 2015, 15, 251. [Google Scholar] [CrossRef]
- Rubio, B.; Stammitti, L.; Cookson, S.J.; Teyssier, E.; Gallusci, P. Small RNA populations reflect the complex dialogue established between heterograft partners in grapevine. Hortic. Res. 2022, 9, uhab067. [Google Scholar] [CrossRef] [PubMed]
- Cookson, S.J.; Ollat, N. Grafting with rootstocks induces extensive transcriptional re-programming in the shoot apical meristem of grapevine. BMC Plant Biol. 2013, 13, 147. [Google Scholar] [CrossRef]
- Chitarra, W.; Perrone, I.; Avanzato, C.G.; Minio, A.; Boccacci, P.; Santini, D.; Gilardi, G.; Siciliano, I.; Gullino, M.L.; Delledonne, M.; et al. Grapevine grafting: Scion transcript profiling and defense-related metabolites induced by rootstocks. Front. Plant Sci. 2017, 8, 654. [Google Scholar] [CrossRef]
- Boyko, A.; Kathiria, P.; Zemp, F.J.; Yao, Y.; Pogribny, I.; Kovalchuk, I. Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (Virus-induced plant genome instability). Nucleic Acids Res. 2007, 35, 1714–1725. [Google Scholar] [CrossRef]
- Deleris, A.; Halter, T.; Navarro, L.D.N.A. DNA methylation and demethylation in plant immunity. Annu. Rev. Phytopathol. 2016, 54, 579–603. [Google Scholar] [CrossRef]
- Dal Bosco, D.; Sinski, I.; Ritschel, P.S.; Camargo, U.A.; Fajardo, T.V.; Harakava, R.; Quecini, V. Expression of disease resistance in genetically modified grapevines correlates with the contents of viral sequences in the T-DNA and global genome methylation. Transgenic Res. 2018, 27, 379–396. [Google Scholar] [CrossRef]
- López Sánchez, A.; Stassen, J.H.; Furci, L.; Smith, L.M.; Ton, J. The role of DNA (de) methylation in immune responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, V.; Daddiego, L.; Cardone, M.F.; Perrella, G.; Sousa, L.; Santos, R.B.; Malhó, R.; Bergamini, C.; Marsico, A.D.; Figueiredo, A.; et al. Transcriptomic and methylation analysis of susceptible and tolerant grapevine genotypes following Plasmopara viticola infection. Physiol. Plant. 2022, 174, 13771. [Google Scholar] [CrossRef]
- Luna, E.; Ton, J. The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal. Behav. 2012, 7, 615–618. [Google Scholar] [CrossRef]
- Weng, K.; Li, Z.Q.; Liu, R.Q.; Wang, L.; Wang, Y.J.; Xu, Y. Transcriptome of Erysiphe necator-infected Vitis pseudoreticulata leaves provides insight into grapevine resistance to powdery mildew. Hortic. Res. 2014, 1, 14049. [Google Scholar] [CrossRef]
- Wang, L.; Ahmad, B.; Liang, C.; Shi, X.; Sun, R.; Zhang, S.; Du, G. Bioinformatics and expression analysis of histone modification genes in grapevine predict their involvement in seed development, powdery mildew resistance, and hormonal signalling. BMC Plant Biol. 2020, 20, 412. [Google Scholar] [CrossRef]
- Agudelo-Romero, P.; Erban, A.; Rego, C.; Carbonell-Bejerano, P.; Nascimento, T.; Sousa, L.; Fortes, A.M. Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J. Exp. Bot. 2015, 66, 1769–1785. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Sandrini, M.; Gambino, G.; Chitarra, W. Double-stranded RNAs (dsRNAs) as a sustainable tool against gray mold (Botrytis cinerea) in grapevine: Effectiveness of different application methods in an open-air environment. Biomolecules 2020, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M. RNA-directed DNA methylation. In Plant Gene Silencing; Matzke, M.A., Matzke, A.J.M., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 83–100. [Google Scholar]
- Gohlke, J.; Scholz, C.J.; Kneitz, S.; Weber, D.; Fuchs, J.; Hedrich, R.; Deeken, R. DNA methylation mediated control of gene expression is critical for development of crown gall tumors. PLoS Genet. 2013, 9, 1003267. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef]
- Pagay, V.; Furlan, T.S.; Kidman, C.M.; Nagahatenna, D. Long-term drought adaptation of unirrigated grapevines (Vitis vinifera L.). Theor. Exp. Plant Physiol. 2022, 34, 215–225. [Google Scholar] [CrossRef]
- Jež-Krebelj, A.; Rupnik-Cigoj, M.; Stele, M.; Chersicola, M.; Pompe-Novak, M.; Sivilotti, P. The physiological impact of GFLV virus infection on grapevine water status: First observations. Plants 2022, 11, 161. [Google Scholar] [CrossRef]
- Nerva, L.; Guaschino, M.; Pagliarani, C.; De Rosso, M.; Lovisolo, C.; Chitarra, W. Spray-induced gene silencing targeting a glutathione S-transferase gene improves resilience to drought in grapevine. Plant Cell Environ. 2022, 45, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, G.; Briglia, N.; Lopez, L.; Amato, D.; Panara, F.; Petrozza, A.; Nuzzo, V. A synthetic cytokinin primes photosynthetic and growth response in grapevine under ion-independent salinity stress. J. Plant Interact. 2022, 17, 789–800. [Google Scholar] [CrossRef]
- Vanden Broeck, A.; Cox, K.; Brys, R.; Castiglione, S.; Cicatelli, A.; Guarino, F.; Vander Mijnsbrugge, K. Variability in DNA methylation and generational plasticity in the Lombardy poplar, a single genotype worldwide distributed since the eighteenth century. Front. Plant Sci. 2018, 9, 1635. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Beckers, G.J.; Langenbach, C.J.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Reimer-Michalski, E.M.; Conrath, U. Innate immune memory in plants. In Seminars in Immunology; Netea, M.G., Ed.; Academic Press: Aachen, Germany, 2016; Volume 28, pp. 319–327. [Google Scholar]
- Gourbal, B.; Pinaud, S.; Beckers, G.J.; Van Der Meer, J.W.; Conrath, U.; Netea, M.G. Innate immune memory: An evolutionary perspective. In Immunological Reviews; Wiley Online Library: Hoboken, NJ, USA, 2018; Volume 283, pp. 21–40. [Google Scholar]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.S.; Duan, C.G. Epigenetic regulation of plant immunity: From chromatin codes to plant disease resistance. aBIOTECH 2023, 4, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Jaskiewicz, M.; Conrath, U.; Peterhänsel, C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011, 12, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Kauss, H.; Jeblick, W. Pretreatment of parsley suspension cultures with salicylic acid enhances spontaneous and elicited production of H2O2. Plant Physiol. 1995, 108, 1171–1178. [Google Scholar] [CrossRef]
- Frost, C.J.; Mescher, M.C.; Carlson, J.E.; De Moraes, C.M. Plant defense priming against herbivores: Getting ready for a different battle. Plant Physiol. 2008, 146, 818–824. [Google Scholar] [CrossRef]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Hamiduzzaman, M.M. B [beta]-Aminobutyric Acid-Induced Resistance in Grapevine Against Downy Mildew (Plasmopara viticola). Ph.D. Thesis, Université de Neuchâtel, Neuchâtel, Switzerland, 2020. [Google Scholar]
- Caffarra, A.; Rinaldi, M.; Eccel, E.; Rossi, V.; Pertot, I. Modelling the impact of climate change on the interaction between grapevine and its pests and pathogens: European grapevine moth and powdery mildew. Agric Ecosyst. Environ. 2012, 148, 89–101. [Google Scholar] [CrossRef]
- Duchêne, E.; Schneider, C. Grapevine and climatic changes: A glance at the situation in Alsace. Agron. Sustain. Dev. 2005, 25, 93–99. [Google Scholar] [CrossRef]
- Fabres, P.J.; Collins, C.; Cavagnaro, T.R.; Rodríguez López, C.M. A concise review on multi-omics data integration for terroir analysis in Vitis vinifera. Front. Plant Sci. 2017, 8, 1065. [Google Scholar] [CrossRef]



| Genes/Proteins | Stress or Function | Epigenetic Process | Organ | V. vinifera cv./ Vitis sp. | Geographic Location | Reference |
|---|---|---|---|---|---|---|
| C2-domain ABA-related (CAR) proteins | Drought, ABA pathway | DNA methylation | Berries | Shiraz | Barossa Valley, Australia | [44] |
| Histone-lysine methyltransferase-related genes (e.g., SUV3) | Heat-drought | Histone modification | Leaves | Cabernet Sauvignon | South Australia | [45] |
| Chitinase-related genes | Cold stress | H3K27me3 | Leaves | V. amurensis | Jilin, China | [46] |
| G-type lectin s-receptor-like serine/threonine-protein kinase | Cold stress | H3K27me3 | Leaves | V. amurensis | Jilin, China | [46] |
| Glucosyltransferases (GTFs) | Cold stress | H3K27me3 | Leaves | V. amurensis | Jilin, China | [46] |
| Peroxidases (PODs) | Cold stress | H3K27me3 | Leaves | V. amurensis | Jilin, China | [46] |
| NAC domain (NAM, ATAF1/2 and CUC2) | Cold stress | H3K27me3 | Leaves | V. amurensis | Jilin, China | [46] |
| Ethylene-insensitive 3 (EIN3) transcription factor | Dormancy/ bud break | DNA methylation | Buds | Kyoho | Nanjing, China | [47] |
| Gibberellin-related (GA) genes | Dormancy/ bud break | DNA demethylation | Buds | Kyoho | Nanjing, China | [47] |
| WRKY domain transcription factors | Cold stress | H3K27me3/DNA demethylation | Leaves | Fleurtai, UD 31-103 | Northern Italy | [47] |
| DEMETER-like DNA demethylase genes (VvDEM1, VvDEM2, VvDEM3) | Dormancy/bud break | DNA demethylation | Buds | Kyoho, V. amurensis | China | [48] |
| MYB (myeloblastosis) domain transcription factors | Cold/drought | miRNAs/DNA demethylation | Leaves | Kyoho, Muscat Hamburg | China | [47,49] |
| bHLH (basic helix–loop–helix) domain transcription factors | Cold/drought/ ABA pathway | miRNAs/DNA demethylation | Leaves | Kyoho, Muscat Hamburg | Beijing, China | [47,49] |
| bZIP (basic-leucine zipper) domain transcription factors | Cold stress | miRNAs | Leaves | Muscat Hamburg | Beijing, China | [49] |
| AP2/ERF (APETALA2/ Ethylene-responsive) transcription factors | Cold stress | miRNAs | Leaves | Muscat Hamburg | Beijing, China | [49] |
| SBP (SQUAMOSA PROMOTER BINDING PROTEIN) transcription factors | Cold stress | miRNAs | Leaves | Muscat Hamburg | Beijing, China | [49] |
| Genes/Proteins | Function | Epigenetic Process | V. vinifera cv./ Vitis sp. | Geographic Location | Reference |
|---|---|---|---|---|---|
| E3 ubiquitin protein ligases | Development–ripening | DNA methylation | Malbec | Gualtallary, Argentina | [58] |
| Pentatricopeptide repeat proteins | Development–ripening | DNA methylation | Malbec | Mendoza, Argentina | [50] |
| F-box protein domain encoding genes | Development–ripening | DNA methylation | Malbec | Mendoza, Argentina | [50] |
| Oxygenase encoding gene (VIT_15s0048g01960) | Sugar content control | DNA methylation | Malbec | Mendoza, Argentina | [50] |
| Oxysterol-binding protein-related protein 4B-like encoding gene (VIT_11s0103g00530) | Sugar content control | DNA methylation | Malbec | Mendoza, Argentina | [50] |
| O-acyltransferase (WSD1-LIKE) gene family | Berry skin dry weight control | DNA methylation | Malbec | Mendoza, Argentina | [50] |
| AP2/ERF (APETALA2/ Ethylene-responsive factor) transcription factors | Berry skin dry weight control | DNA methylation | Malbec | Mendoza, Argentina | [50] |
| Stilbene synthase VaSTS10 | Resveratrol biosynthesis | DNA methylation | Shiraz | Barossa Valley, Australia | [60] |
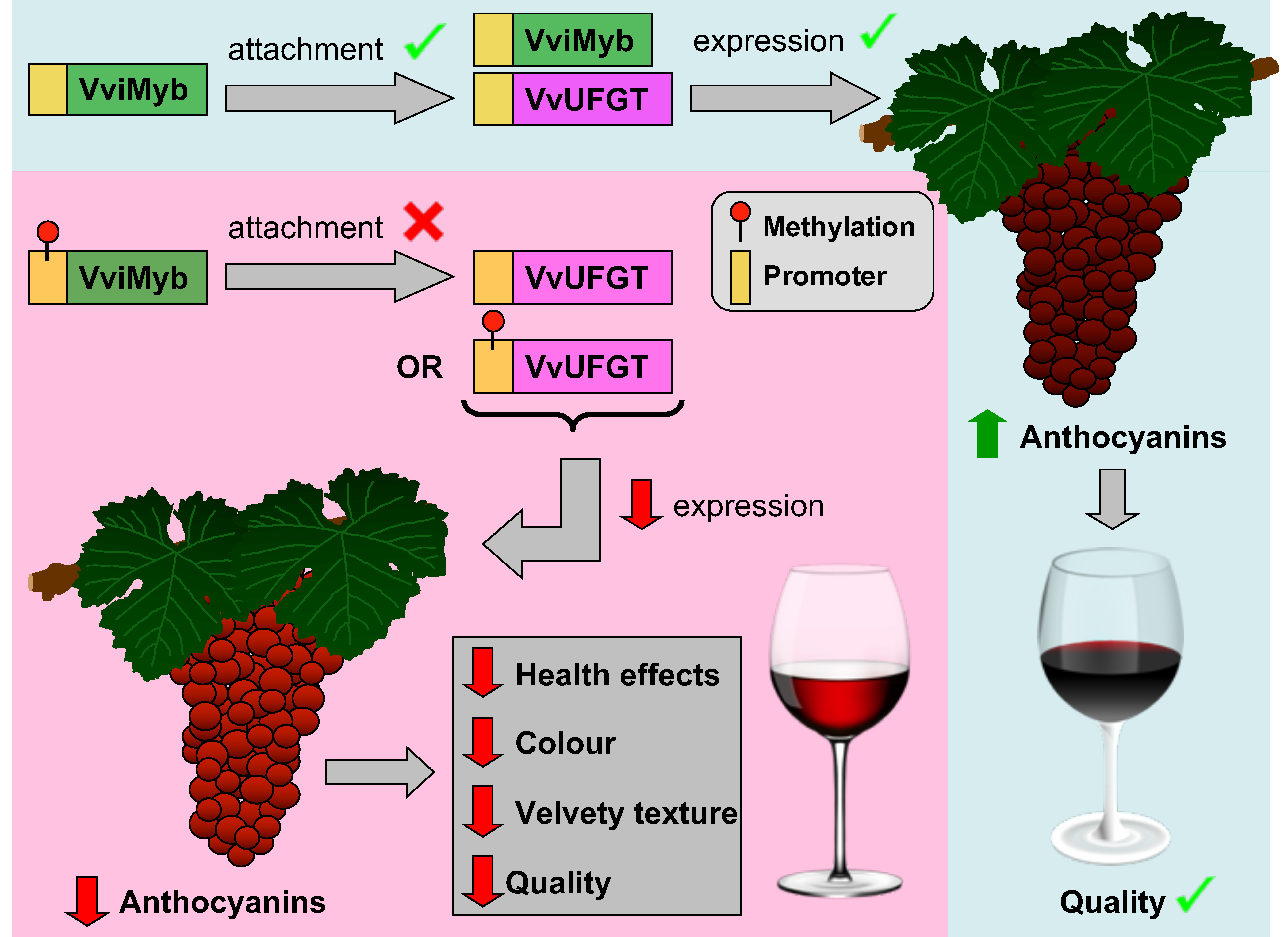
| UDP-glucose-flavonoid 3-O-glucosyltransferase VvUFGT | Anthocyanin biosynthesis | DNA methylation | Gamay Teinturier | Bordeaux, France | [67] |
| SET DOMAIN GROUP (SDG) proteins | Flowering/ grape development | H3K27me3 | Cabernet Sauvignon | Central Chile | [68] |
| No apical meristem (NAM) gene | Ripening control | H3K27me3 | Pinot noir | Hong Kong, China | [69] |
| MADS-box transcription factors | Ripening control | H3K27me3 | Pinot noir | Hong Kong, China | [69] |
| VvMYBA1, VvMYBA2 | Anthocyanin biosynthesis | DNA methylation | Gamay Teinturier | BarossaValley, Australia | [67] |
| VvMYB114 | Flavonoid biosynthesis | miRNAs | Dilkhush, Bangalore Blue, Red Globe | Bengaluru, India | [70] |
| VvO-methyltransferase 3 (VvOMT3) | Methoxypyrazines (MPs) biosynthesis | H3K27me3 | V. amurensis | Jilin, China | [46] |
| Dihydroflavonol reductase VvDFR | Anthocyanin accumulation | DNA methylation | Kyoho | Nanjing, China | [47] |
| Glutathione S-transferase VvGST | Anthocyanin accumulation | DNA methylation | Kyoho | Nanjing, China | [47] |
| chalcone synthase VvCHS | Anthocyanin accumulation | DNA methylation | Kyoho | Nanjing, China | [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venios, X.; Gkizi, D.; Nisiotou, A.; Korkas, E.; Tjamos, S.E.; Zamioudis, C.; Banilas, G. Emerging Roles of Epigenetics in Grapevine and Winegrowing. Plants 2024, 13, 515. https://doi.org/10.3390/plants13040515
Venios X, Gkizi D, Nisiotou A, Korkas E, Tjamos SE, Zamioudis C, Banilas G. Emerging Roles of Epigenetics in Grapevine and Winegrowing. Plants. 2024; 13(4):515. https://doi.org/10.3390/plants13040515
Chicago/Turabian StyleVenios, Xenophon, Danai Gkizi, Aspasia Nisiotou, Elias Korkas, Sotirios E. Tjamos, Christos Zamioudis, and Georgios Banilas. 2024. "Emerging Roles of Epigenetics in Grapevine and Winegrowing" Plants 13, no. 4: 515. https://doi.org/10.3390/plants13040515






