Genetic Variation and Heritability for Hydrogen Cyanide in Fresh Cassava Roots: Implications for Low-Cyanide Cassava Breeding
Abstract
:1. Introduction
2. Results
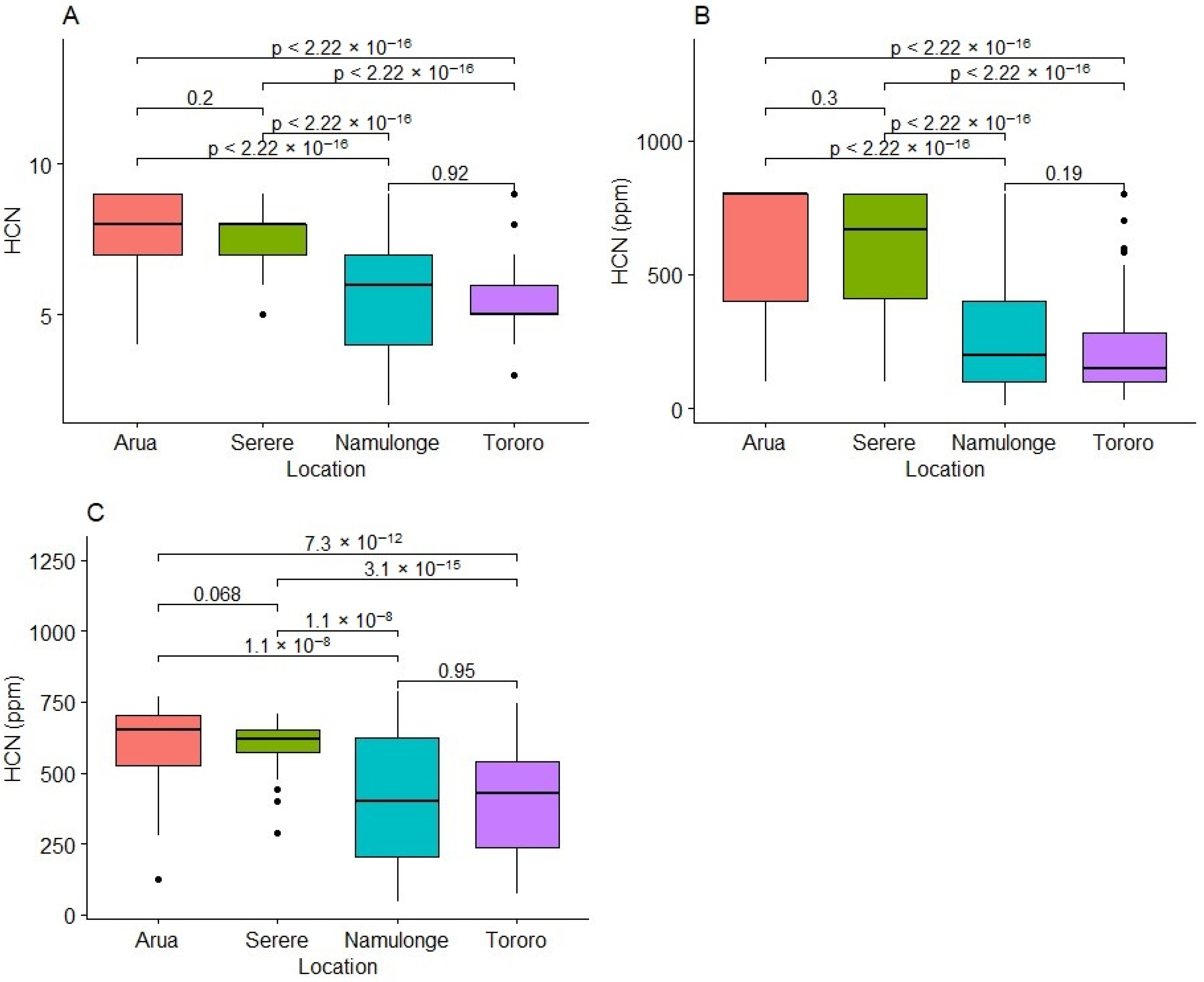
2.1. Variability for HCN in Fresh Cassava Roots
2.2. Accession Ranking for Fresh Cassava Root HCN Content Using BLUPS
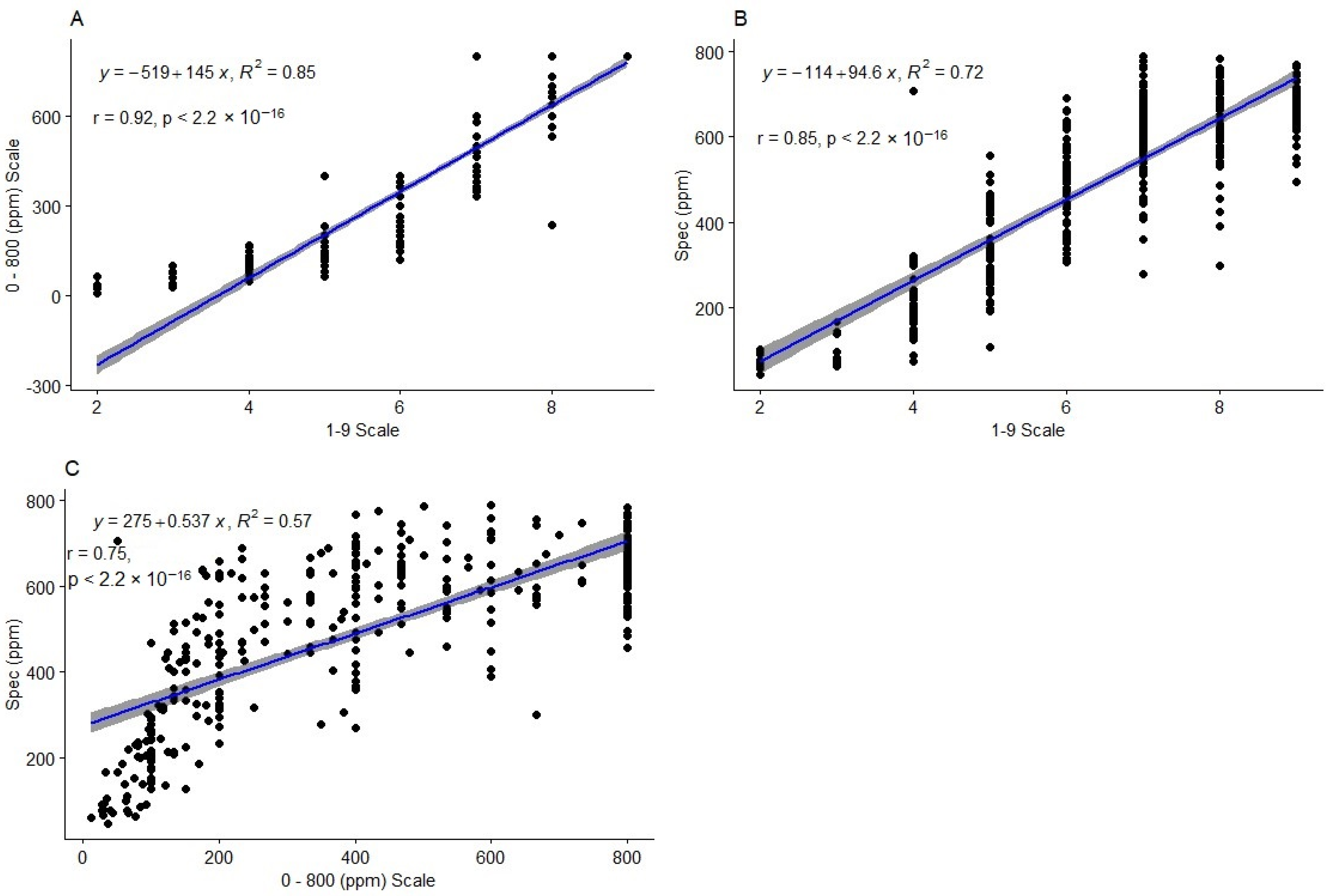
2.3. Comparison of Phenotyping Method Performance
2.3.1. Correlation and Regression between the Methods
2.3.2. Comparison of Residuals
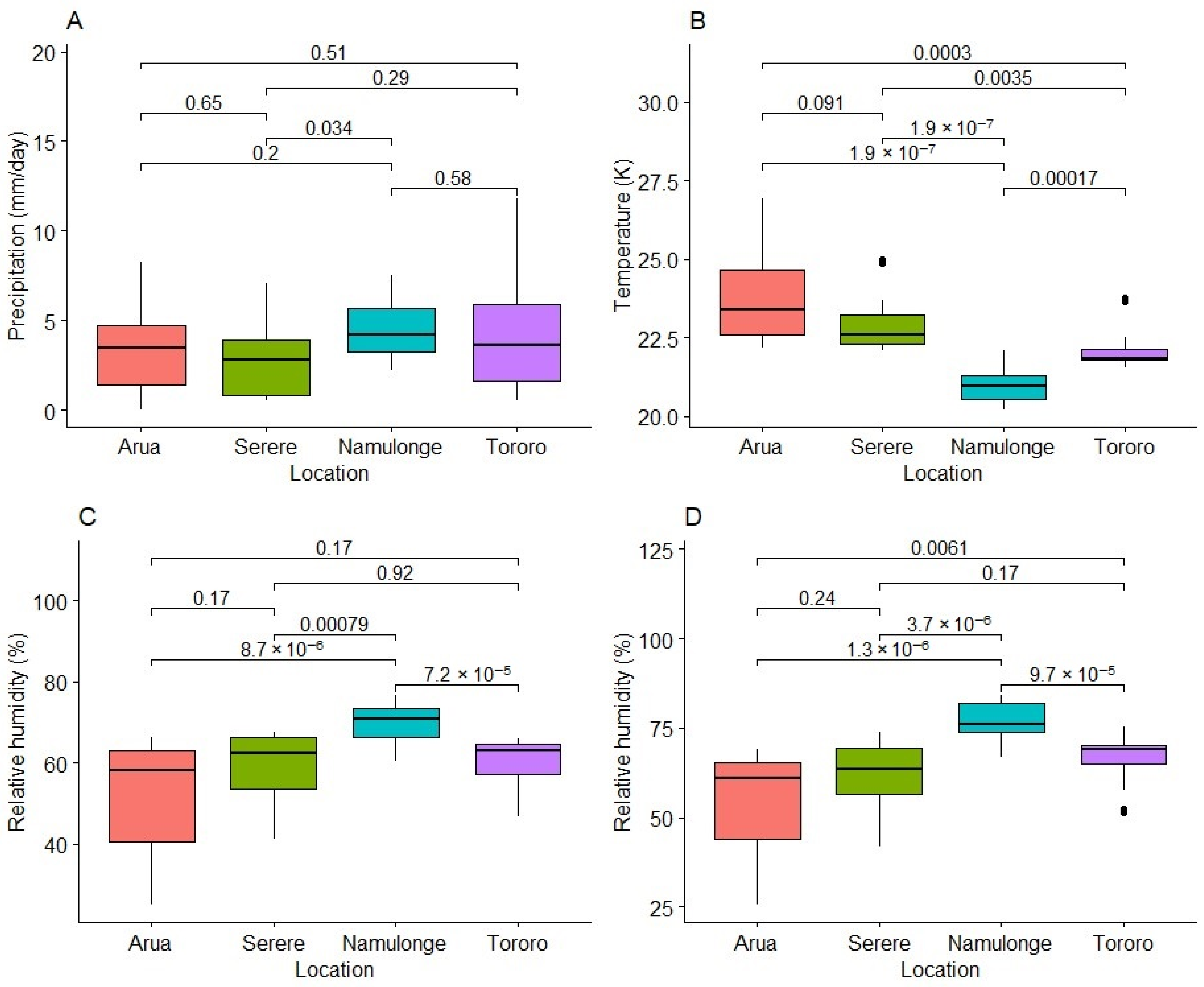
2.4. Variation in Weather Variables at Experimental Test Locations
3. Discussion
3.1. Genetic Variability and Heritability for HCN
3.2. Effect of the Environment on Fresh Root HCN Content
3.3. Accuracy of HCN Phenotyping Methods
4. Materials and Methods
4.1. Description of the Study Area
4.2. Description of Study Materials and Field Trial Establishment
4.3. Sample Selection, Preparation, and Data Collection
4.3.1. Sample Selection
4.3.2. Sample Preparation
4.4. Data Collection
4.4.1. Measurement of HCN
4.4.2. Weather Data
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manyong, V.M.; Makinde, K.O.; Bokanga, M.; Whyte, J. The Contribution of IITA-Improved Cassava to Food Security in Sub-Saharan Africa: An Impact Study; IITA: Ibadan, Nigeria, 2000; p. 4. [Google Scholar]
- El-Sharkawy, M.A. Stress-Tolerant Cassava: The Role of Integrative Ecophysiology-Breeding Research in Crop Improvement. Open J. Soil Sci. 2012, 2, 162–186. [Google Scholar] [CrossRef]
- Iragaba, P.; Hamba, S.; Nuwamanya, E.; Kanaabi, M.; Nanyonjo, R.A.; Mpamire, D.; Muhumuza, N.; Khakasa, E.; Tufan, H.A.; Kawuki, R.S. Identification of cassava quality attributes preferred by Ugandan users along the food chain. Int. J. Food Sci. Technol. 2021, 56, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Nuwamanya, E.; Kawuki, R.S. Quantification of starch physicochemical characteristics in a cassava segregating population. Afr. Crop Sci. J. 2008, 16, 191–202. [Google Scholar] [CrossRef]
- Nanyonjo, A.R.; Dufour, D.; Kawuki, R.S.; Kyazze, F.; Esuma, W.; Wembabazi, E.; Nuwamanya, E.; Tufan, H. Assessment of end user traits and physicochemical qualities of cassava flour: A case of Zombo district, Uganda. Int. J. Food Sci. Technol. 2021, 56, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.R.; Bradbury, J.H. Total cyanide determination of plants and foods using the picrate and acid hydrolysis methods. Food Chem. 2002, 77, 107–114. [Google Scholar] [CrossRef]
- Daniel, L.; Da, J.; Francisco, C.; Zelder, F.; Bergenståhl, B.; Dejmek, P. Straightforward rapid spectrophotometric quantification of total cyanogenic glycosides in fresh and processed cassava products. Food Chem. 2014, 158, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.V.; Yeoh, H.H.; Bradbury, J.H. Simple picrate paper kit for determination of the cyanogenic potential of cassava flour. J. Sci. Food Agric. 1998, 76, 39–48. [Google Scholar] [CrossRef]
- Zagrobelny, M.; Møller, B.L. Cyanogenic glucosides in the biological warfare between plants and insects: The Burnet moth-Birdsfoot trefoil model system. Phytochemistry 2011, 72, 1585–1592. [Google Scholar] [CrossRef]
- Gleadow, R.M.; Møller, B.L. Cyanogenic glycosides: Synthesis, physiology, and phenotypic plasticity. Annu. Rev. Plant Biol. 2014, 65, 155–185. [Google Scholar] [CrossRef]
- Bradbury, J.H.; Egan, S.V. Rapid screening assay of cyanide content of cassava. Phytochem. Anal. 1992, 3, 91–94. [Google Scholar] [CrossRef]
- McKey, D.; Cavagnaro, T.R.; Cliff, J.; Gleadow, R. Chemical ecology in coupled human and natural systems: People, manioc, multitrophic interactions and global change. Chemoecology 2010, 20, 109–133. [Google Scholar] [CrossRef]
- Wheatley, C.C.; Ghuzel, G.; Zakhia, N. The Nature of the Tuber. In Encylopedia of Food Sciences and Nutrition, 2nd ed.; Cabellero, B., Ed.; Academic Press: Cambridge, MA, USA, 2003; pp. 964–969. [Google Scholar]
- FAO/WHO. Book Review: Safety Evaluation of Certain Food Additives and Contaminants. Nutr. Health 2001, 15, 74. [Google Scholar] [CrossRef]
- Akintonwa, A.; Tunwashe, O.L. Fatal Cyanide Poisoning from Cassava-based Meal. Hum. Exp. Toxicol. 1992, 11, 47–49. [Google Scholar] [CrossRef] [PubMed]
- Alitubeera, P.H.; Eyu, P.K.; Benon, A.; Alex, R.; Zhu, B. Outbreak of Cyanide Poisoning Caused by Consumption of Cassava Flour. Morb. Mortal. Wkly. Rep. 2019, 68, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Nzwalo, H.; Cliff, J. Konzo: From Poverty, Cassava, and Cyanogen Intake to Toxico-Nutritional Neurological Disease. PLoS Negl. Trop. Dis. 2011, 5, e1051. [Google Scholar] [CrossRef] [PubMed]
- Cliff, J.; Muquingue, H.; Nhassico, D.; Nzwalo, H.; Bradbury, J.H. Konzo and continuing cyanide intoxication from cassava in Mozambique. Food Chem. Toxicol. 2011, 49, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Nhassico, D.; Muquingue, H.; Cliff, J.; Cumbana, A.; Bradbury, J.H. Rising African cassava production, diseases due to high cyanide intake and control measures. J. Sci. Food Agric. 2008, 2049, 2043–2049. [Google Scholar] [CrossRef]
- Nakabonge, G.; Samukoya, C.; Baguma, Y. Local varieties of cassava: Conservation, cultivation and use in Uganda. Environ. Dev. Sustain. 2018, 20, 2427–2445. [Google Scholar] [CrossRef] [PubMed]
- Bechoff, A.; Tomlins, K.; Fliedel, G.; Becerra Lopez-lavalle, L.A.; Westby, A.; Hershey, C.; Dufour, D. Cassava traits and end-user preference: Relating traits to consumer liking, sensory perception, and genetics. Crit. Rev. Food Sci. Nutr. 2018, 58, 547–567. [Google Scholar] [CrossRef]
- Dominique, D.; Clair, H.; Hamaker, R.B.; Lorenzen, J. Integrating end-user preferences into breeding programmes for roots tubers and bananas. Int. J. Food Sci. Technol. 2021, 56, 1071–1075. [Google Scholar] [CrossRef]
- Thiele, G.; Dufour, D.; Vernier, P.; Mwanga, R.O.M.; Parker, M.L.; Schulte, G.E.; Teeken, B.; Wossen, T.; Gotor, E.; Kikulwe, E.; et al. A review of varietal change in roots, tubers and bananas: Consumer preferences and other drivers of adoption and implications for breeding. Int. J. Food Sci. Technol. 2021, 56, 1076–1092. [Google Scholar] [CrossRef]
- Polar, V.; Ashby, J.A.; Thiele, G.; Tufan, H. When is choice empowering? Examining gender differences in varietal adoption through case studies from sub-saharan africa. Sustainability 2021, 13, 3678. [Google Scholar] [CrossRef]
- Takam Tchuente, H.N.; Fongang Fouepe, G.H.; Mbwentchou Yao, D.C.; Mathe, S.; Teeken, B. Varietal diversity as a lever for cassava variety development: Exploring varietal complementarities in Cameroon. J. Sci. Food Agric. 2023. [Google Scholar] [CrossRef]
- Honfozo, L.F.; Djibril Moussa, I.M.; Adinsi, L.; Bouniol, A.; Adetonah, S.; Chadare, F.J.; Padonou, W.S.; Hounhouigan, J.D.; Forsythe, L.; Akissoe, N.H. Cross-approaches for advising cassava trait-preferences for boiling. Cogent Food Agric. 2023, 9, 2253716. [Google Scholar] [CrossRef]
- Bezerra, C.; Ferreira, E.; Cunha, R.L.; Tomé, J.; Neto, D.F.; Silva, R.D.S. Chemical root traits differentiate ‘bitter’ and ‘sweet’ cassava clones from the Amazon. Crop Breed. Appl. Biotechnol. 2019, 19, 77–85. [Google Scholar] [CrossRef]
- Mkumbira, J.; Chiwona-Karltun, L.; Lagercrantz, U.; Mahungu, N.M.; Saka, J.; Mhone, A.; Bokanga, M.; Brimer, L.; Gullberg, U.; Rosling, H. Classification of cassava into “bitter” and “cool” in Malawi: From farmers’ perception to characterisation by molecular markers. Euphytica 2003, 132, 7–22. [Google Scholar] [CrossRef]
- Bello, O.B.; Ige, S.A.; Azeez, M.A.; Afolabi, M.S.; Abdulmalic, S.Y.; Mohamood, J. Heritability and Genetic Advance for Grain Yield and its Component Characters in Maize (Zea mays L.). Int. J. Plant Res. 2012, 2, 138–145. [Google Scholar]
- Imakumbili, M.L.E.; Semu, E.; Semoka, J.M.R.; Abass, A.; Mkamilo, G. Soil nutrient adequacy for optimal cassava growth, implications on cyanogenic glucoside production: A case of konzo-affected Mtwara region, Tanzania. PLoS ONE 2019, 14, e0216708. [Google Scholar] [CrossRef]
- Banea-Mayambu, J.P.; Tylleskär, T.; Gitebo, N.; Matadi, N.; Gebre-Medhin, M.; Rosling, H. Geographical and seasonal association between linamarin and cyanide exposure from cassava and the upper motor neurone disease konzo in former Zaire. Trop. Med. Int. Health 1997, 2, 1143–1151. [Google Scholar] [CrossRef]
- Meredith, G.B.; Sylivia, V.E.; Bradbury, J.H. Picrate paper kits for determination of total cyanogens in cassava roots and all forms of cyanogens in cassava products. J. Sci. Food Agric. 1999, 79, 593–601. [Google Scholar]
- Haque, M.R.; Bradbury, J.H. Preparation of linamarin from cassava leaves for use in a cassava cyanide kit. Food Chem. 2004, 85, 27–29. [Google Scholar] [CrossRef]
- Fukuda, W.M.G.; Guevara, C.L.; Kawuki, R.; Ferguson, M.E. Selected Morphological and Agronomic Descriptors for the Characterization of Cassava; IITA: Ibadan, Nigeria, 2010; p. 19. [Google Scholar] [CrossRef]
- Morales, N.; Ogbonna, A.C.; Ellerbrock, B.J.; Bauchet, G.J.; Tantikanjana, T.; Tecle, I.Y.; Powell, A.F.; Lyon, D.; Menda, N.; Simoes, C.C.; et al. Breedbase: A digital ecosystem for modern plant breeding. G3 2022, 12, jkac078. [Google Scholar] [CrossRef]
- Dufour, D.; Dufour, E.; Tirrone, G.; Escobar, A.; Giraldo, A.; Sanchez, T. Evaluation of highland cassava for starch production in Colombia. In Proceedings of the First Scientific Meeting of the Global Cassava Partnership GCP-1, Cassava Meeting the Challenges of the New Millenium, Ghent, Belgique, 21–25 July 2008; p. 96. [Google Scholar]
- Torres, L.G.; de Oliveira, E.J.; Ogbonna, A.C.; Fonseca, F.; Simiqueli, G.F.; Kantar, M.B. Can Cross-Country Genomic Predictions Be a Reasonable Strategy to Support Germplasm Exchange?—A Case Study with Hydrogen Cyanide in Cassava. Front. Plant Sci. 2021, 12, 742638. [Google Scholar] [CrossRef]
- Ogbonna, A.C.; Braatz de Andrade, L.R.; Rabbi, I.Y.; Mueller, L.A.; Jorge de Oliveira, E.; Bauchet, G.J. Large-scale genome-wide association study, using historical data, identifies conserved genetic architecture of cyanogenic glucoside content in cassava (Manihot esculenta Crantz) root. Plant J. 2021, 105, 754–770. [Google Scholar] [CrossRef]
- Manze, F.; Rubaihayo, P.; Ozimati, A.; Gibson, P.; Esuma, W.; Bua, A.; Alicai, T.; Omongo, C.; Kawuki, R.S. Genetic Gains for Yield and Virus Disease Resistance of Cassava Varieties Developed over the Last Eight Decades in Uganda. Front. Plant Sci. 2021, 12, 651992. [Google Scholar] [CrossRef]
- Schmidt, P.; Hartung, J.; Bennewitz, J.; Hans-Peter, P. Heritability in plant breeding on a genotype-difference basis. Genetics 2019, 212, 991–1008. [Google Scholar] [CrossRef]
- Giovanny, E.C.P. Heritability: Meaning and Computation. 2019. Available online: https://excellenceinbreeding.org/sites/default/files/manual/EiB-M2_Heritability_18-02-20.pdf (accessed on 10 October 2023).
- Bernardo, R. Parental selection, number of breeding populations, and size of each population in inbred development. Theor. Appl. Genet. 2003, 107, 1252–1256. [Google Scholar] [CrossRef]
- Mondo, J.M.; Irenge, A.B.; Ayagirwe, R.B.; Dontsop-Nguezet, P.M.; Karume, K.; Njukwe, E.; Mapatano, S.M.; Zamukulu, P.M.; Basimine, G.C.; Musungayi, E.M.; et al. Determinants of Adoption and Farmers’ Preferences for Cassava Varieties in Kabare Territory, Eastern Democratic Republic of Congo. Am. J. Rural Dev. 2019, 7, 44–52. [Google Scholar] [CrossRef]
- Zhong, Y.; Xu, T.; Ji, S.; Wu, X.; Zhao, T.; Li, S.; Zhang, P.; Li, K.; Lu, B. Effect of ultrasonic pretreatment on eliminating cyanogenic glycosides and hydrogen cyanide in cassava. Ultrason. Sonochem. 2021, 78, 105742. [Google Scholar] [CrossRef]
- Cardoso, A.P.; Mirione, E.; Ernesto, M.; Massaza, F.; Cliff, J.; Rezaul, H.M.; Bradbury, J.H. Processing of cassava roots to remove cyanogens. J. Food Compos. Anal. 2005, 18, 451–460. [Google Scholar] [CrossRef]
- Bandna, C. Effect of processing on the cyanide content of cassava. J. Microbiol. Biotechnol. Food Sci. 2012, 2, 947–958. [Google Scholar]
- Adewusi, S.R.A.; Akindahunsi, A.A. Cassava processing, consumption, and cyanide toxicity. J. Toxicol. Environ. Health 1994, 43, 13–23. [Google Scholar] [CrossRef] [PubMed]
- CCDN. Working Together to Eliminate Cyanide Poisoning, Konzo, Tropic Ataxic Neuropathy (TAN) and Neurolathyrism. 2011, 18, 1–4. Available online: https://biblio.ugent.be/publication/2002992/file/2003018.pdf (accessed on 15 October 2023).
- Nuwamanya, E.; Turyasingura, C.; Magumba, I.; Katungisa, A.; Alicai, T. Cyanogenic Potential Variations within Plot, Plant and Roots of Cassava Varieties Grown in the Same Environment. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2023, 93, 365–372. [Google Scholar] [CrossRef]
- Jørgensen, K.; Bak, S.; Busk, P.K.; Sørensen, C.; Olsen, C.E.; Puonti-Kaerlas, J.; Møller, B.L. Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol. 2005, 139, 363–374. [Google Scholar] [CrossRef]
- Njankouo, N.Y.; Mounjouenpou, P.; Kansci, G.; Kenfack, M.J.; Meguia, F.M.P.; Natacha, N.E.N.S.; Akhobakoh, M.M.; Nyegue, A. Influence of cultivars and processing methods on the cyanide contents of cassava (Manihot esculenta Crantz) and its traditional food products. Sci. Afr. 2019, 5, e00119. [Google Scholar] [CrossRef]
- Bokanga, M.; Ekanayake, I.; Dixon, A.; Porto, M. Genotype-environment interactions for cyanogenic potential in cassava. Acta Hortic. 1994, 375, 131–140. [Google Scholar] [CrossRef]
- Zidenga, T.; Siritunga, D.; Sayre, R.T. Cyanogen metabolism in cassava roots: Impact on protein synthesis and root development. Front. Plant Sci. 2017, 8, 220. [Google Scholar] [CrossRef]
- Almazroui, M.; Saeed, F.; Saeed, S.; Nazrul, I.M.; Ismail, M.; Klutse, N.A.B.; Siddiqui, M.H. Projected Change in Temperature and Precipitation over Africa from CMIP6. Earth Syst. Environ. 2020, 4, 455–475. [Google Scholar] [CrossRef]
- Panter, D.M.; Allen, F.L. Using best linear unbiased predictions to enhance breeding for yield in soybean: II. Selection of superior crosses from a limited number of yield trials. Crop Sci. 1995, 35, 397–405. [Google Scholar] [CrossRef]
- Piepho, H.P.; Möhring, J.; Melchinger, A.E.; Büchse, A. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 2008, 161, 209–228. [Google Scholar] [CrossRef]
- Molenaar, H.; Boehm, R.; Piepho, H.P. Phenotypic selection in ornamental breeding: It’s better to have the BLUPs than to have the BLUEs. Front. Plant Sci. 2018, 9, 1511. [Google Scholar] [CrossRef] [PubMed]
- Namakula, B.F.; Nuwamanya, E.; Kanaabi, M.; Wembambazi, E.; Kawuki, R.S. Predicting starch content of cassava with near infrared spectroscopy in Ugandan cassava germplasm. J. Near Infrared Spectrosc. 2023, 31, 256–262. [Google Scholar] [CrossRef]
- Nkouaya Mbanjo, E.G.; Hershberger, J.; Peteti, P.; Agbona, A.; Ikpan, A.; Ogunpaimo, K.; Kayondo, S.I.; Abioye, R.S.; Nafiu, K.; Alamu, E.O.; et al. Predicting starch content in cassava fresh roots using near-infrared spectroscopy. Front. Plant Sci. 2022, 13, 990250. [Google Scholar] [CrossRef] [PubMed]
- Nuwamanya, E.; Enoch, W.; Kanaabi, M.; Namakula, F.B.; Katungisa, A.; Lyatumi, I.; Esuma, W.; Alamu, E.O.; Dufour, D.; Kawuki, R.; et al. Development and validation of near-infrared spectroscopy procedures for prediction of cassava root dry matter and amylose contents in Ugandan cassava germplasm. J. Sci. Food Agric. 2023. [Google Scholar] [CrossRef] [PubMed]
- Kanaabi, M.; Namakula, F.B.; Nuwamanya, E.; Kayondo, I.S.; Muhumuza, N.; Wembabazi, E.; Iragaba, P.; Nandudu, L.; Nanyonjo, A.R.; Baguma, J.; et al. Rapid analysis of hydrogen cyanide in fresh cassava roots using NIRSand machine learning algorithms: Meeting end user demand for low cyanogenic cassava. Plant Genome 2023. [Google Scholar] [CrossRef] [PubMed]
- Robert, P.; Brault, C.; Rincent, R.; Segura, V. Phenomic Selection: A New and Efficient Alternative to Genomic Selection. In Genomic Prediction of Complex Traits. Methods in Molecular Biology; Ahmadi, N., Bartholomé, J., Eds.; Humana: New York, NY, USA, 2022; Volume 2467. [Google Scholar] [CrossRef]
- Siritunga, D.; Sayre, R.T. Generation of cyanogen-free transgenic cassava. Planta 2003, 217, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Magambo, S.; Nabatanzi, A.; Alicai, T.; Wembabazi, E.; Oketcho, K.; Nakalembe, I.; Wagaba, H. Somatic embryo production and GFP genetic transformation in elite Ugandan cassava genotypes. Sci. Afr. 2024, 23, e02039. [Google Scholar] [CrossRef]
- Taylor, N.; Gaitán-Solís, E.; Moll, T.; Trauterman, B.; Jones, T.; Pranjal, A.; Trembley, C.; Abernathy, V.; Corbin, D.; Fauquet, C.M. A High-throughput Platform for the Production and Analysis of Transgenic Cassava (Manihot esculenta) Plants. Trop Plant Biol. 2012, 5, 127–139. [Google Scholar] [CrossRef]
- Ozimati, A.; Kawuki, R.; Esuma, W.; Kayondo, I.S.; Wolfe, M. Training Population Optimization for Prediction of Cassava Brown Streak Disease Resistance in West African Clones. G3 2018, 8, 3903–3913. [Google Scholar] [CrossRef]
- Buontempo, C.; Thépaut, J.N.; Bergeron, C. Copernicus Climate Change Service. IOP Conf. Ser. Earth Environ. Sci. 2020, 509, 10–12. [Google Scholar] [CrossRef]
- Brown, D.; de Sousa, K.; van Etten, J. ag5Tools: An R package for downloading and extracting agrometeorological data from the AgERA5 database. SoftwareX 2023, 21, 101267. [Google Scholar] [CrossRef]
- Boogaard, H.; Schubert, J.; De Wit, A.; Lazebnik, J.; Hutjes, R.; Van der Grijn, G. Agrometeorological Indicators from 1979 to Present Derived from Reanalysis. Copernicus Climate Change Service (C3S) Climate Data Store (CDS). 2020. Available online: https://cds.climate.copernicus.eu/cdsapp#!/dataset/10.24381/cds.6c68c9bb?tab=overview (accessed on 15 October 2023).

| Mean Square | ||||
|---|---|---|---|---|
| Source of Variation | Degrees of Freedom | 1–9_Scale | HCN_0–800_Scale | HCN_Spec |
| Clone | 63 | 5.069 *** | 130,338 *** | 57,706 *** |
| Location | 3 | 127.548 *** | 3,384,185 *** | 1,127,633 *** |
| Replication | 1 | 1.133 | 33,359 | 670,202 *** |
| Accession: Location | 149 | 1.571 ** | 33,731 * | 27,349 *** |
| Residuals | 147 | 1.085 | 27,283 | 14,280 |
| Location | HCN_1–9_Scale | SD | H2 | HCN_0–800_Scale | SD | H2 | HCN_Spec | SD | H2 |
|---|---|---|---|---|---|---|---|---|---|
| Arua | 7.70 a | 1.26 | 0.46 | 614.24 a | 231.72 | 0.64 | 601.85 a | 136.03 | 0.50 |
| Serere | 7.58 a | 0.97 | 0.54 | 595.68 a | 205.842 | 0.24 | 608.19 a | 66.16 | 0.22 |
| Namulonge | 5.45 b | 1.77 | 0.50 | 277.48 b | 216.6 | 0.52 | 407.0 b | 231.15 | 0.56 |
| Tororo | 5.75 b | 1.34 | 0.48 | 263.03 b | 204.29 | 0.56 | 399.39 b | 181.92 | 0.40 |
| CV (%) | 15.6 | 38.50 | 24.60 | ||||||
| Vg | 0.48 | 13,990 | 3596 | ||||||
| Vge | 0.35 | 4943 | 7748 | ||||||
| VRes | 1.04 | 24,897 | 14,093 | ||||||
| H2 | 0.26 | 0.32 | 0.14 |
| 1–9 Scale | 0–800 ppm Scale | HCN_Spec | ||||||
|---|---|---|---|---|---|---|---|---|
| Accession | BLUP | Classification | Accession | BLUP | Classification | Accession | BLUP | Classification |
| TONGOLO | 1.306 | High | TONGOLO | 257.776 | High | KAZIMWENGE | 63.186 | High |
| NYAMATIA | 1.085 | High | NYAMATIA | 225.384 | High | UG16F300P068 | 59.250 | High |
| KAZIMWENGE | 0.883 | High | QUININE | 181.413 | High | TONGOLO | 56.682 | High |
| QUININE | 0.810 | High | KAZIMWENGE | 177.962 | High | UG16F158P004 | 55.176 | High |
| UG16F303P006 | 0.785 | High | UG16F303P006 | 124.465 | High | UG16F063P006 | 54.696 | High |
| UG16F290P332 | 0.768 | High | UG16F063P006 | 120.565 | High | NYAMATIA | 53.768 | High |
| UG16F303P009 | 0.758 | High | UG16F303P009 | 114.728 | High | UG16F294P011 | 44.224 | High |
| UG16F158P004 | 0.734 | High | UG16F158P004 | 103.961 | High | UG16F293P151 | 42.927 | High |
| UG16F294P011 | 0.707 | High | UG16F300P068 | 102.232 | High | UG16F077P002 | 42.798 | High |
| UG16F063P006 | 0.672 | High | UG16F159P001 | 91.997 | High | QUININE | 36.430 | High |
| EDWARAT | −0.628 | Low | UG16F290P073 | −102.508 | Low | EDWARAT | −51.843 | Low |
| UG16F290P128 | −0.743 | Low | UG16F290P040 | −107.260 | Low | UG16F290P073 | −52.257 | Low |
| UG16F293P082 | −0.753 | Low | UG16F314P005 | −107.724 | Low | UG16F290P128 | −52.338 | Low |
| UG16F303P005 | −0.779 | Low | UG110017 | −109.308 | Low | UG16F293P066 | −52.881 | Low |
| UG16F290P295 | −0.882 | Low | UG16F303P005 | −119.431 | Low | UG16F293P169 | −54.483 | Low |
| UG16F293P169 | −1.039 | Low | UG16F057P001 | −122.378 | Low | UG16F293P082 | −56.428 | Low |
| UG16F057P001 | −1.090 | Low | UG16F290P295 | −125.102 | Low | UG16F057P001 | −70.226 | Low |
| UG16F001P013 | −1.262 | Low | UG16F293P169 | −137.138 | Low | UG16F001P013 | −71.706 | Low |
| UG16F293P066 | −1.361 | Low | UG16F001P013 | −189.676 | Low | UG16F290P295 | −81.562 | Low |
| UG16F290P075 | −1.574 | Low | UG16F290P075 | −226.642 | Low | UG16F290P075 | −101.391 | Low |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanaabi, M.; Settumba, M.B.; Nuwamanya, E.; Muhumuza, N.; Iragaba, P.; Ozimati, A.; Namakula, F.B.; Kayondo, I.S.; Baguma, J.K.; Nanyonjo, A.R.; et al. Genetic Variation and Heritability for Hydrogen Cyanide in Fresh Cassava Roots: Implications for Low-Cyanide Cassava Breeding. Plants 2024, 13, 1186. https://doi.org/10.3390/plants13091186
Kanaabi M, Settumba MB, Nuwamanya E, Muhumuza N, Iragaba P, Ozimati A, Namakula FB, Kayondo IS, Baguma JK, Nanyonjo AR, et al. Genetic Variation and Heritability for Hydrogen Cyanide in Fresh Cassava Roots: Implications for Low-Cyanide Cassava Breeding. Plants. 2024; 13(9):1186. https://doi.org/10.3390/plants13091186
Chicago/Turabian StyleKanaabi, Michael, Mukasa B. Settumba, Ephraim Nuwamanya, Nicholas Muhumuza, Paula Iragaba, Alfred Ozimati, Fatumah B. Namakula, Ismail S. Kayondo, Julius K. Baguma, Ann Ritah Nanyonjo, and et al. 2024. "Genetic Variation and Heritability for Hydrogen Cyanide in Fresh Cassava Roots: Implications for Low-Cyanide Cassava Breeding" Plants 13, no. 9: 1186. https://doi.org/10.3390/plants13091186






