PiSCP1 and PiCDPK2 Localize to Peroxisomes and Are Involved in Pollen Tube Growth in Petunia Inflata
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of a cDNA Clone Product That Interacts with the PiCDPK1 Kinase Domain
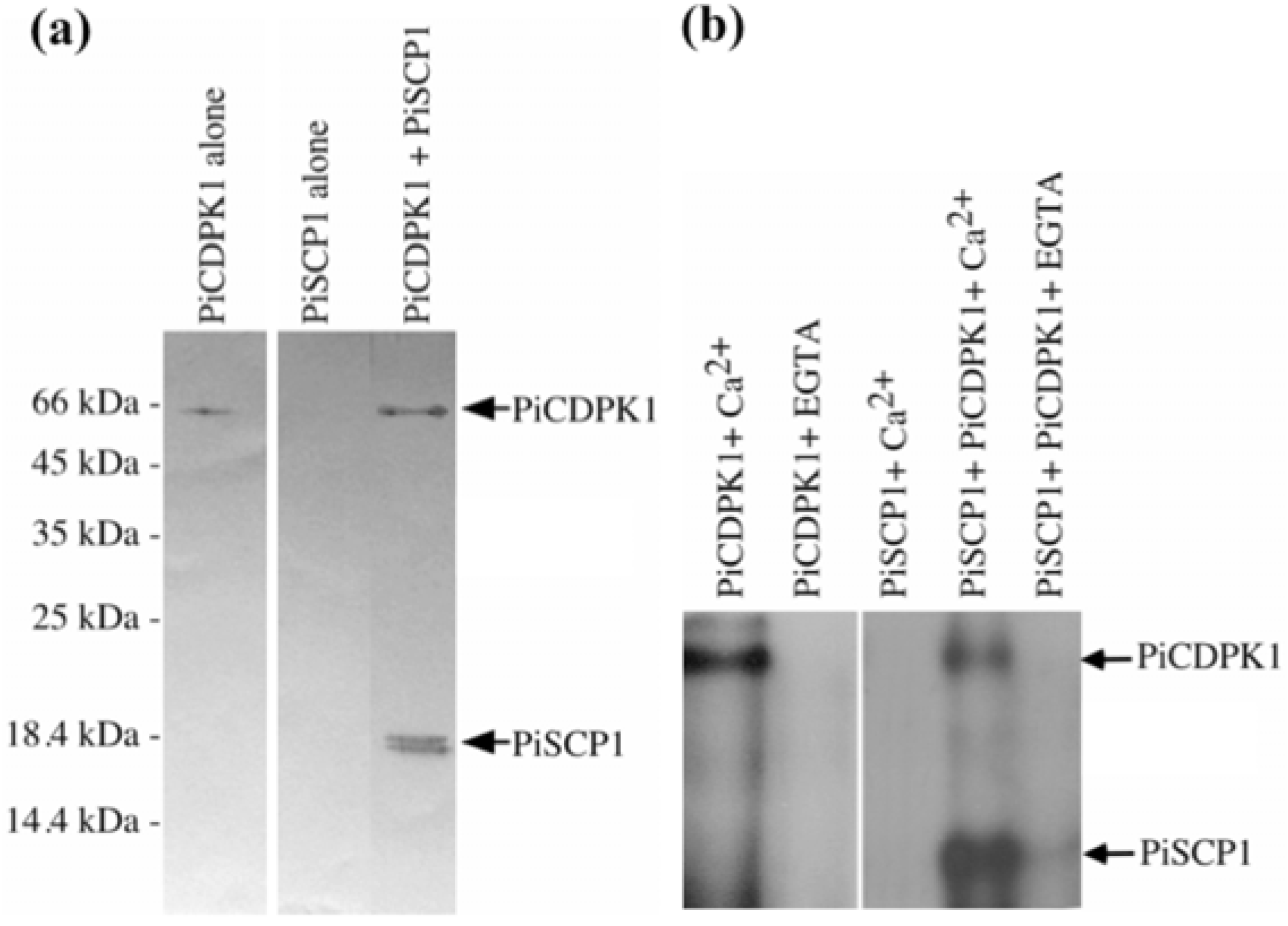
2.2. PiCDPK1 Interacts with and Phosphorylates PiSCP1 in Vitro

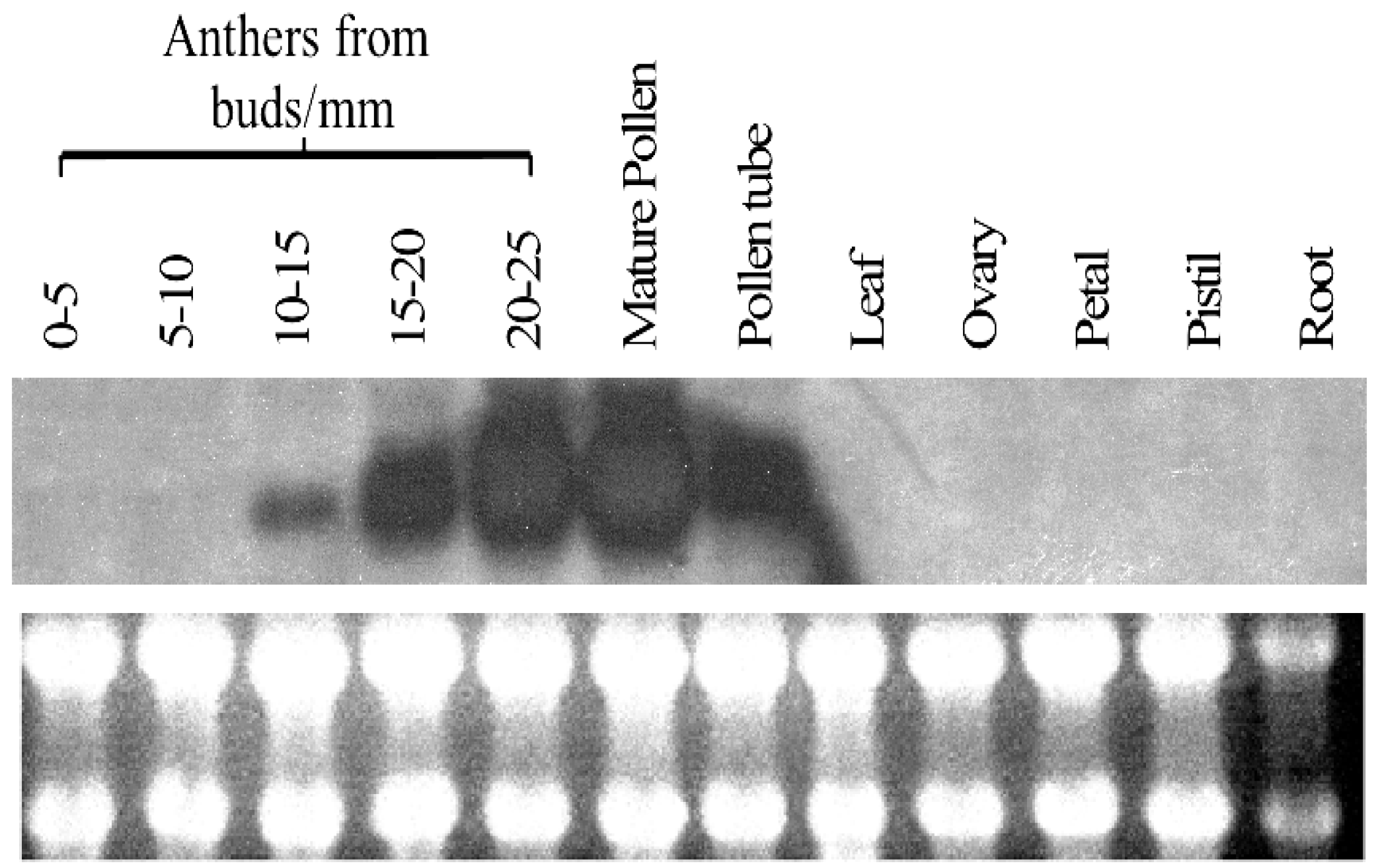
2.3. Expression Profile of PiSCP1
2.4. Transient Over-Expression of PiSCP1 in Pollen Tubes
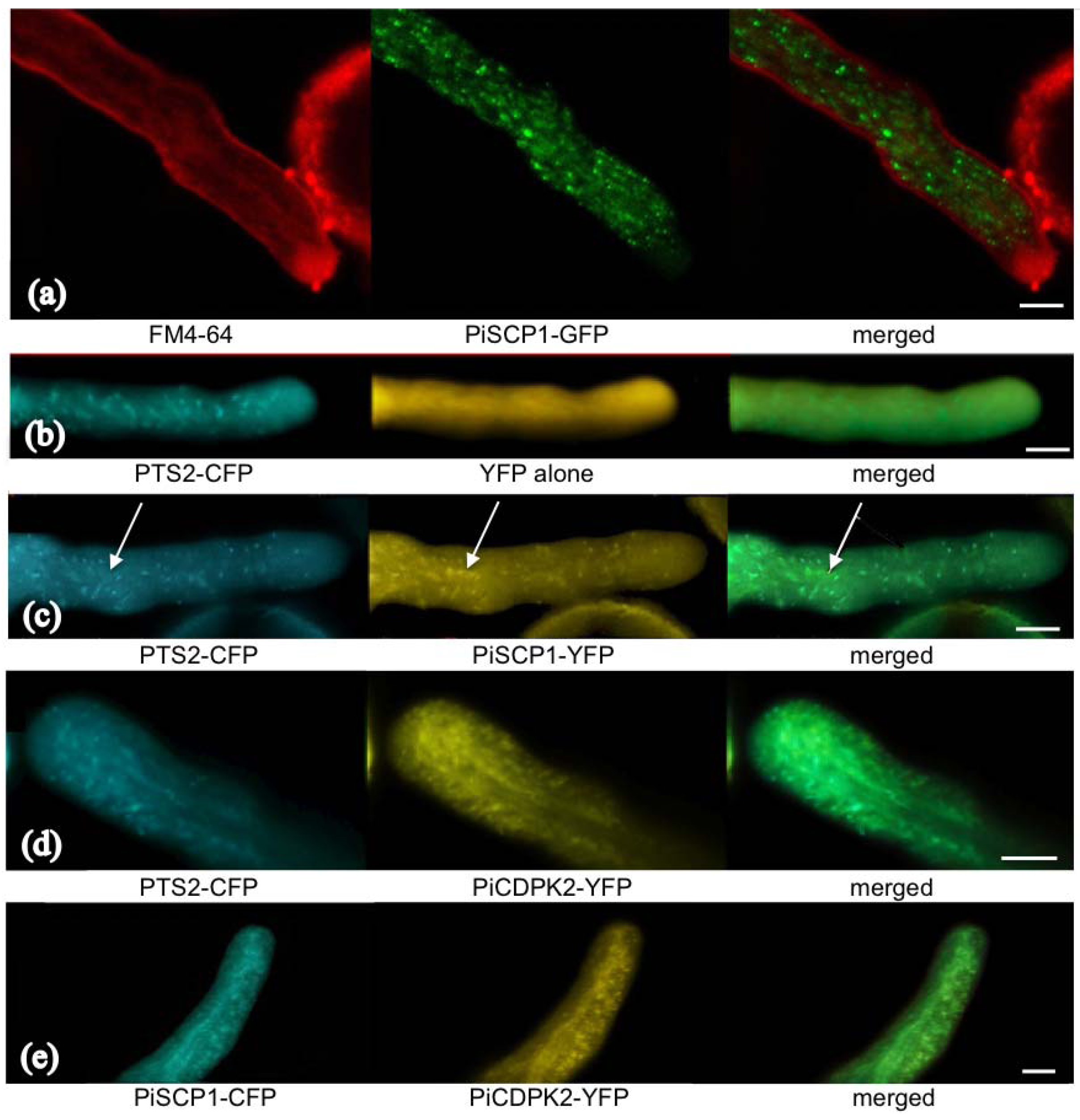
2.5. PiCDPK2 Localized to Peroxisomes and Co-Localized with PiSCP1
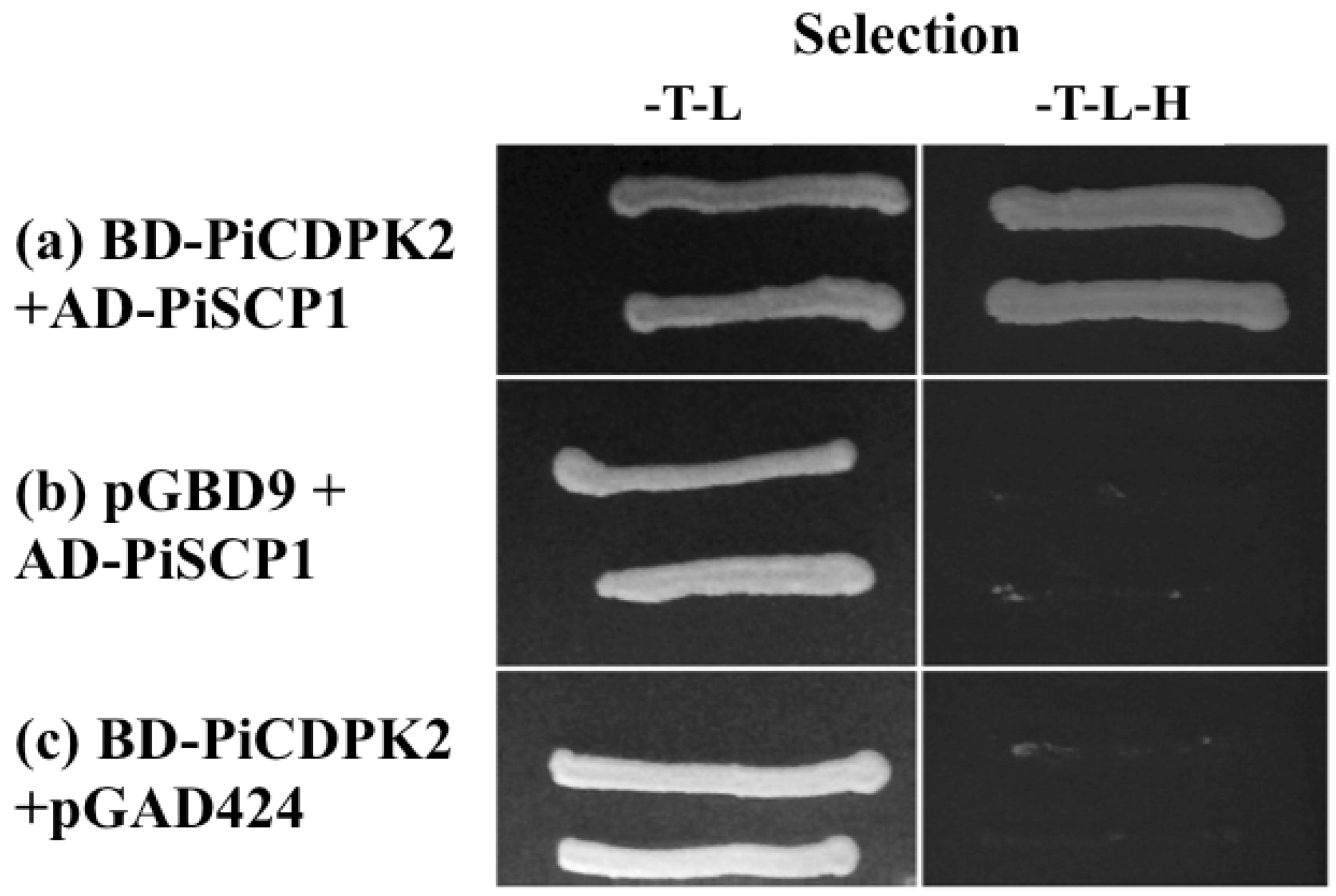
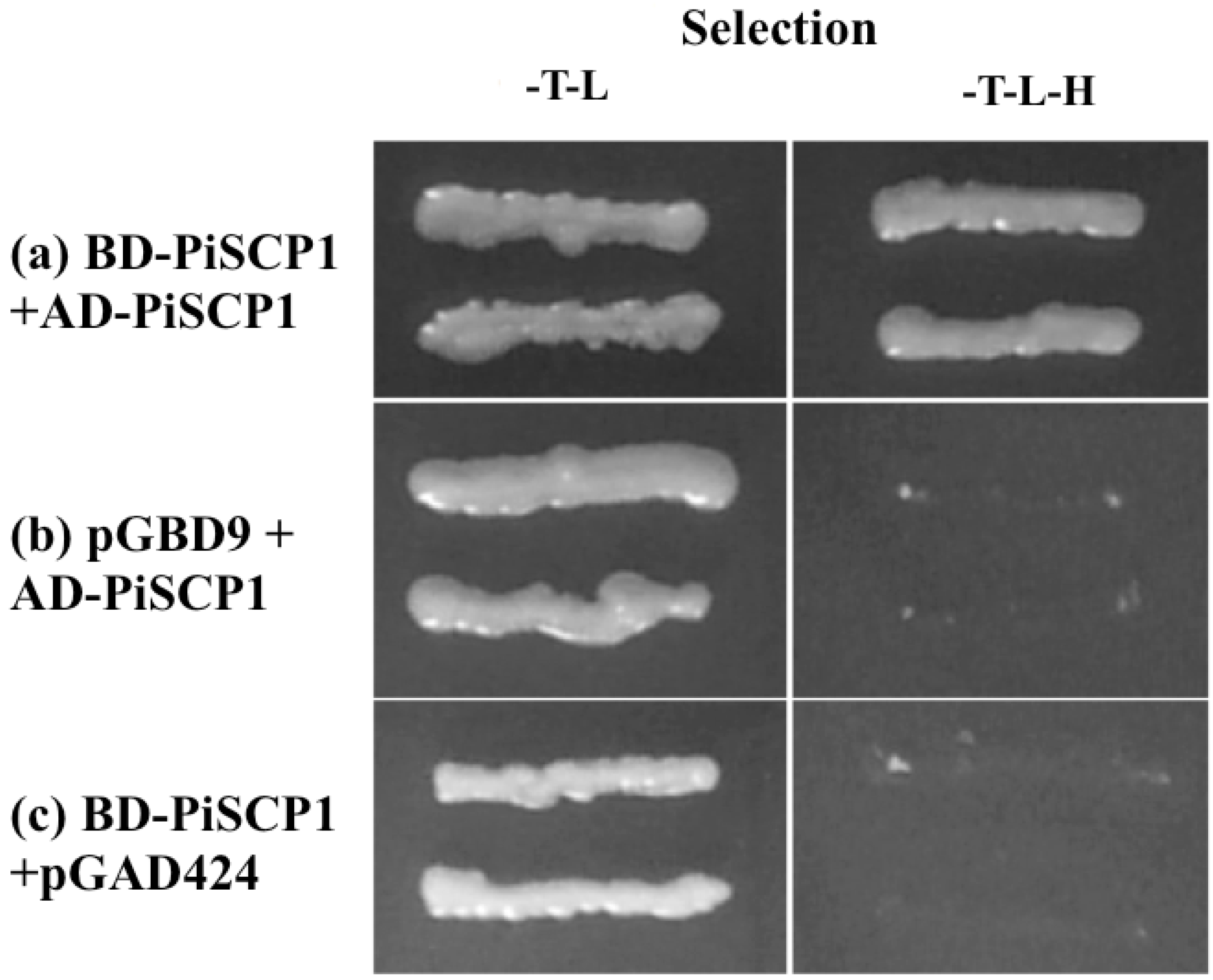
2.6. PiSCP1 Interacts with PiCDPK2 in the Yeast 2 Hybrid System
2.7. PiSCP1 Forms Multimers
3. Experimental Section
3.1. Plant Material
3.2. Yeast Two Hybrid Library Screening
3.3. Isolation of Full-Length PiSCP1 cDNA
3.4. RNA Gel Blot Analysis
3.5. Plasmid Constructs for Recombinant Proteins
3.6. Expression and Purification of PiCDPK1 and PiSCP1
3.7. In Vitro Biding Assay and in Vitro Phosphorylation Assay
3.8. Transient Expression in Pollen
3.9. Analysis of Transformed Pollen Tubes
4. Conclusions
Acknowledgments
References and Notes
- Miller, D.D.; Callaham, D.A.; Gross, D.J.; Hepler, P.K. Free Ca2+ gradient in growing pollen tubes of Lilium. J. Cell. Sci. 1992, 101, 7–12. [Google Scholar]
- Pierson, E.S.; Miller, D.D.; Callaham, D.A.; Shipley, A.M.; Rivers, B.A.; Cresti, M.; Hepler, P.K. Pollen tube growth is coupled to the extracellular calcium ion flux and the intracellular calcium gradient: Effect of BAPTA-type buffers and hypertonic media. Plant Cell 1994, 6, 1815–1828. [Google Scholar]
- Mahló, R.; Trewavas, A. Localized apical increases of cytosolic free calcium control pollen tube orientation. Plant Cell 1996, 8, 1935–1949. [Google Scholar]
- Messerli, M.; Robinson, K.R. Tip localized Ca2+ pulses are coincident with peak pulsatile growth rates in pollen tubes of Lilium longiflorum. J. Cell. Sci. 1997, 110, 1269–1278. [Google Scholar]
- Rathore, K.S.; Cork, R.J.; Robinson, K.R. A cytoplasmic gradient of Ca2+ is correlated with the growth of lily pollen tubes. Dev. Biol. 1991, 148, 612–619. [Google Scholar] [CrossRef]
- Lin, Y.K.; Wang, Y.L.; Zhu, J.-K.; Yang, Z.B. Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell 1996, 8, 293–303. [Google Scholar]
- Hepler, P.K.; Kunkel, J.G.; Rounds, C.M.; Winship, L.J. Calcium entry into pollen tubes. Trends Plant Sci. 2012, 17, 32–8. [Google Scholar] [CrossRef]
- Steinhorst, L.; Kudla, J. Calcium—A central regulator of pollen germination and tube growth. Biochim. Biophys. Acta 2012, in press. [Google Scholar]
- Lin, Y.; Yang, Z. Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip-growth. Plant Cell 1997, 9, 1647–1659. [Google Scholar]
- Kost, B.; Lemichez, P.; Sielhofer, P.; Hong, Y.; Tolias, K.; Carpenter, C.; Chua, N.H. Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 1999, 145, 317–330. [Google Scholar] [CrossRef]
- Li, H.; Lin, Y.; Heath, R.M.; Zhu, M.X.; Yang, Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 1999, 11, 1731–1742. [Google Scholar]
- Zheng, Z.L.; Yang, Z. The Rop GTPase switch turns on polar growth in pollen. Trends Plant Sci. 2000, 5, 298–303. [Google Scholar] [CrossRef]
- Qin, Y.; Yang, Z. Rapid tip growth: Insights from pollen tubes. Semin. Cell Dev. Biol. 2011, 22, 816–824. [Google Scholar] [CrossRef]
- Gu, Y.; Fu, Y.; Dowd, P.; Li, S.; Vernoud, V.; Gilroy, S.; Yang, Z. A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell. Biol. 2005, 169, 127–38. [Google Scholar] [CrossRef]
- Yoon, G.M.; Dowd, P.E.; Gilroy, S.; McCubbin, A.G. Calcium-Dependent Protein Kinase isoforms have distinct functions in pollen tube growth, including regulating polarity. Plant Cell 2006, 18, 867–878. [Google Scholar] [CrossRef]
- Myers, C.; Romanowsky, S.M.; Barron, Y.D.; Garg, S.; Azuse, C.L.; Curran, A.; Davis, R.M.; Hatton, J.; Harmon, A.C.; Harper, J.F. Calcium-Dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009, 59, 528–539. [Google Scholar] [CrossRef]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Pical, C.; Fredlund, K.M.; Petit, P.X.; Sommarin, M.; Møller, I.M. The outer membrane of plant mitochondria contains a calcium-dependent protein kinase and multiple phosphoproteins. FEBS Lett. 1993, 336, 347–351. [Google Scholar] [CrossRef]
- Patharkar, O.R.; Cushman, J.C. A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J. 2000, 24, 679–691. [Google Scholar] [CrossRef]
- Lu, S.X.; Hrabak, E.M. An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol. 2002, 128, 1008–1021. [Google Scholar] [CrossRef]
- Anil, V.S.; Harmon, A.C.; Rao, K.S. Temporal association of Ca(2+)-dependent protein kinase with oil bodies during seed development in Santalum album L.: Its biochemical characterization and significance. Plant Cell Physiol. 2003, 44, 367–376. [Google Scholar]
- Dammann, C.; Ichida, A.; Hong, B.; Romanowsky, S.M.; Hrabak, E.M.; Harmon, A.C.; Pickard, B.G.; Harper, J.F. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 2003, 132, 1840–1848. [Google Scholar] [CrossRef]
- Anil, V.S.; Sankara Rao, K. Purification and characterization of a Ca2+-dependent protein kinase from sandalwood (Santalum album L.): Evidence for Ca2+-induced conformational changes. Phytochemistry 2001, 58, 203–212. [Google Scholar]
- Cheng, S.; Willmann, M.R.; Chen, H.; Sheen, J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar]
- Harper, J.F.; Harmon, A. Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 2005, 6, 55–566. [Google Scholar]
- Pina, C.; Pinto, F.; Feijó, J.A.; Becker, J.D. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005, 138, 744–756. [Google Scholar] [CrossRef]
- Battey, N.; Venis, M.A. Separation of calmodulin from calcium-activated protein kinase by using calcium dependent hydrophobic interaction chromatography. Anal. Biochem. 1988, 170, 116–122. [Google Scholar] [CrossRef]
- Johnson, T.L.; Olsen, L.J. Import of the peroxisomal targeting signal type 2 protein 3-ketoacyl-Coenzyme A thiolase into glyoxysomes. Plant Physiol. 2003, 133, 1991–1999. [Google Scholar] [CrossRef]
- Glover, J.R.; Andrews, D.W.; Rachubinski, R.A. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. USA 1994, 91, 10541–10545. [Google Scholar] [CrossRef]
- Prado, A.M.; Porterfield, D.M.; Feijó, J.A. Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 2004, 131, 2707–2714. [Google Scholar] [CrossRef]
- Yoon, G.M. Investigation into the function of calmodulin-like domain protein kinase mediated signaling pathways in pollen tube growth. Ph.D. Thesis, Washington State University, Pullman WA, USA, 2006. [Google Scholar]
- Ito, T.; Nakata, M.; Fukazawa, J.; Ishida, S.; Takahashi, Y. Alteration of substrate specificity: The variable N-terminal domain of tobacco Ca2+-Dependent Protein Kinase is important for substrate recognition. Plant Cell 2010, 22, 1592–1604. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Frietsch, S.; Kim, T.; Dixon, M.B.; Schroeder, J.I. The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr. Biol. 2008, 18, 63–68. [Google Scholar] [CrossRef]
- Hu, X.; Neill, S.J.; Tang, Z.; Cai, W. Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiol. 2005, 137, 663–670. [Google Scholar] [CrossRef]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Sandalio, L.M.; Valderrama, R.; Palma, J.M.; Lupiáñez, J.A.; del Río, L.A. Localization of nitric oxide in plant peroxisomes. J. Biol. Chem. 1999, 274, 36729–36733. [Google Scholar]
- Del Río, L.A.; Corpas, F.J.; Sandalio, L.M.; Palma, J.M.; Gómez, M.; Barroso, J.B. Reactive oxygen species, antioxidant species and nitric oxide in peroxisomes. J. Exp. Bot. 2002, 53, 1255–1272. [Google Scholar] [CrossRef]
- Prado, A.M.; Colaço, R.; Moreno, N.; Silva, A.C.; Feijó, J.A. Targeting of pollen tubes to ovules is dependent on Nitric Oxide (NO) signaling. Mol. Plant 2008, 1, 703–714. [Google Scholar] [CrossRef]
- McInnis, S.M.; Desikan, R.; Hancock, J.T.; Hiscock, S.J. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: Potential signaling crosstalk? New Phytol. 2006, 172, 221–228. [Google Scholar] [CrossRef]
- Stael, S.; Wurzinger, B.; Mair, A.; Mehlmer, N.; Vothknecht, U.C.; Teige, M. Plant organellar calcium signalling: An emerging field. J. Exp. Bot. 2012, 63, 1525–1542. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B.W. Arabidopsis chloroplast chaperonin 10 is a calmodulin-binding protein. Biochem. Biophys. Res. Commun. 2000, 275, 601–607. [Google Scholar] [CrossRef]
- Coca, M.; San Segundo, B. AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 2010, 63, 526–540. [Google Scholar] [CrossRef]
- James, P.; Halladay, J.; Craig, E.A. Genomic libraries and a host strain designated for highly efficient two-hybrid selection in yeast. Genetics 1996, 144, 1425–1436. [Google Scholar]
- Gietz, R.D.; Schiestl, R.H.; Willems, A.R.; Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. [Google Scholar] [CrossRef]
- Skirpan, A.L.; McCubbin, A.G.; Ishimizu, T.; Wang, X.; Hu, Y.; Dowd, P.E.; Ma, H.; Kao, T.-H. Isolation and characterization of kinase interacting protein 1, a pollen protein that interacts with the kinase domain of PRK1, receptor-like kinase of Petunia. Plant Physiol. 2001, 126, 1480–1492. [Google Scholar] [CrossRef]
- Parton, R.M.; Fischer-Parton, S.; Watahiki, M.K.; Trewavas, A.J. Dynamics of the apical vesicle accumulation and the rate of growth are related in individual pollen tubes. J. Cell Sci. 2001, 114, 2685–2695. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guo, F.; Yoon, G.M.; McCubbin, A.G. PiSCP1 and PiCDPK2 Localize to Peroxisomes and Are Involved in Pollen Tube Growth in Petunia Inflata. Plants 2013, 2, 72-86. https://doi.org/10.3390/plants2010072
Guo F, Yoon GM, McCubbin AG. PiSCP1 and PiCDPK2 Localize to Peroxisomes and Are Involved in Pollen Tube Growth in Petunia Inflata. Plants. 2013; 2(1):72-86. https://doi.org/10.3390/plants2010072
Chicago/Turabian StyleGuo, Feng, Gyeong Mee Yoon, and Andrew G. McCubbin. 2013. "PiSCP1 and PiCDPK2 Localize to Peroxisomes and Are Involved in Pollen Tube Growth in Petunia Inflata" Plants 2, no. 1: 72-86. https://doi.org/10.3390/plants2010072



